Identification of Redeye, a new sleep-regulating protein whose expression is modulated by sleep amount
Abstract
In this study, we report a new protein involved in the homeostatic regulation of sleep in Drosophila. We conducted a forward genetic screen of chemically mutagenized flies to identify short-sleeping mutants and found one, redeye (rye) that shows a severe reduction of sleep length. Cloning of rye reveals that it encodes a nicotinic acetylcholine receptor α subunit required for Drosophila sleep. Levels of RYE oscillate in light–dark cycles and peak at times of daily sleep. Cycling of RYE is independent of a functional circadian clock, but rather depends upon the sleep homeostat, as protein levels are up-regulated in short-sleeping mutants and also in wild type animals following sleep deprivation. We propose that the homeostatic drive to sleep increases levels of RYE, which responds to this drive by promoting sleep.
https://doi.org/10.7554/eLife.01473.001eLife digest
Almost all animals need to sleep, including most insects. In experiments in the 1980s, a group of rats that were completely deprived of sleep died within only a few weeks. Sleep has been implicated in processes including tissue repair, memory consolidation and, more recently, the removal of waste materials from the brain. However, a full understanding of why we sleep is still lacking.
As anyone who has experienced jetlag can testify, the timing of the sleep/wake cycle is governed by the circadian clock, which leads us to feel sleepy at certain points of the day–night cycle and alert at others. The duration of sleep is regulated by a second process called sleep/wake homeostasis. The longer we remain awake, the more the body’s need for sleep—or ‘sleep drive’—increases, until it becomes almost impossible to stay awake any longer. Whereas many components of the circadian clock have been identified, relatively little is known about the molecular basis of this second process.
Now, Shi et al. have identified a key component of the sleep/wake homeostatic system using the fruit fly and genetic model organism, Drosophila. Flies with a mutation in one particular gene, subsequently named redeye, were found to sleep only half as long as normal flies. While the insects were able to fall asleep, they would wake again only a few minutes later.
Redeye encodes a subunit of a receptor that has previously been implicated in the control of wakefulness, known as the nicotinic acetylcholine receptor. Mutant flies had normal circadian rhythms, suggesting that their sleep problems were the result of disrupted sleep/wake homeostasis. Consistent with this, levels of redeye showed two daily peaks, one corresponding to night-time sleep and the second to the time at which flies would normally take an afternoon siesta. This suggests that redeye signals an acute need for sleep, and then helps to maintain sleep once it is underway.
While redeye is not thought to be the factor that triggers sleep per se, it is directly under control of the sleep homeostat, and may be a useful biomarker for sleep deprivation. The fact that redeye was identified in fruit flies, a species whose genome has been fully sequenced, opens up the possibility of further studies to identify the genetic basis of sleep regulation.
https://doi.org/10.7554/eLife.01473.002Introduction
Sleep is a common and prominent behavior in almost all vertebrate animals and also in most invertebrates (Cirelli, 2009; Sehgal and Mignot, 2011). The function of sleep is a mystery, but it is surely of great importance to animals, as prolonged sleep deprivation can lead to death. Anatomical studies in mammals and birds have revealed brain structures and neurotransmitters that regulate sleep and wakefulness (Saper et al., 2010). However, our understanding of the molecular mechanisms that drive the need to sleep is still in its infancy, partially due to the challenge of performing genetic experiments with mammalian models. In the past decade, several premier genetic organisms have been introduced into the sleep field, including fruit flies, worms and zebra fish. Mammalian counterparts of some sleep components identified in these model animals also regulate sleep (Joho et al., 2006), which argues that (i) behavioral genetics in lower organisms provides an efficient tool to identify sleep components, (ii) at least some of the mechanisms underlying sleep are conserved through evolution.
A two-process model for sleep regulation has been widely accepted by the sleep field. Process C (the circadian clock) controls the timing, in other words the onset and offset of sleep, whereas process S (the sleep homeostat) regulates sleep duration based on the sleep pressure built up during prior wakefulness (Borbely, 1982). This simple model explains sleep related phenomena, including sleep rebound after sleep deprivation. Molecular mechanisms of circadian control have been well characterized (Zheng and Sehgal, 2012), but, as noted above, relatively little is known about process S. Forward genetic screens in Drosophila have led to the identification of several mutants with altered sleep length, but while the genes implicated by these mutants are required for implementation of sleep drive, they have not yet been directly linked to this drive (Sehgal and Mignot, 2011).
Using a forward genetic screen, we identified a new sleep mutant we termed redeye (rye), which is directly controlled by the homeostatic drive to sleep. The rye mutation maps to a nicotinic acetylcholine receptor (nAChR), which interacts with a previously identified sleep-regulating protein, SLEEPLESS (SSS). Levels of RYE are expressed cyclically, in conjunction with the sleep state, and reflect sleep need such that they are up-regulated after sleep deprivation and in short-sleeping mutants. We conclude that homeostatic sleep drive promotes sleep at least in part by increasing expression of RYE.
Results
Identification of redeye, a short-sleeping mutant
As previous screens have identified sleep components on the X chromosome and the second chromosome (Cirelli et al., 2005; Koh et al., 2008; Stavropoulos and Young, 2011), we sought to identify sleep-altering mutations on the third chromosome in Drosophila. To this end, we generated a stock of iso31 (Ryder et al., 2004) flies carrying a newly isogenized third chromosome and treated males of this stock with 10–25 mM ethyl-methanesulfonate (EMS). Individual male progeny were bred to achieve homozygosity of the third chromosome, and females of the F3 generation were tested for daily sleep patterns in the presence of light:dark cycles (Koh et al., 2008). As seen in previous screens (Koh et al., 2008), sleep amounts were normally distributed in the 1857 lines assayed (Figure 1A, Figure 1—source data 1). Two homozygous mutant lines showed a severe reduction of sleep length, and we focused on one of these that we named redeye (rye). rye homozygotes show a >50% reduction in both daytime and night-time sleep (Figure 1C). The activity (average number of beam crossings during periods of activity) of rye mutants is comparable with that of wild type controls, suggesting it is not a hyperactive mutant (Figure 1B). rye heterozygotes have slightly less sleep than wild type, suggesting that the rye mutation is partially dominant (Figure 1B,C). The dramatic reduction of baseline sleep results largely from a shortening of the average sleep episode duration (Figure 1D), which is indicative of a defect in sleep maintenance. The average number of sleep episodes at night is actually increased significantly in rye mutants, perhaps because the sleep homeostat senses sleep loss and compensates by initiating more bouts. We also assayed rye mutants in sleep deprivation assays and found that they did not lose much additional sleep in response to mechanical stimulation (data not shown). We surmise that an inability to sustain sleep bouts leads to the buildup of high sleep need in rye mutants, which makes them resist any further sleep loss.
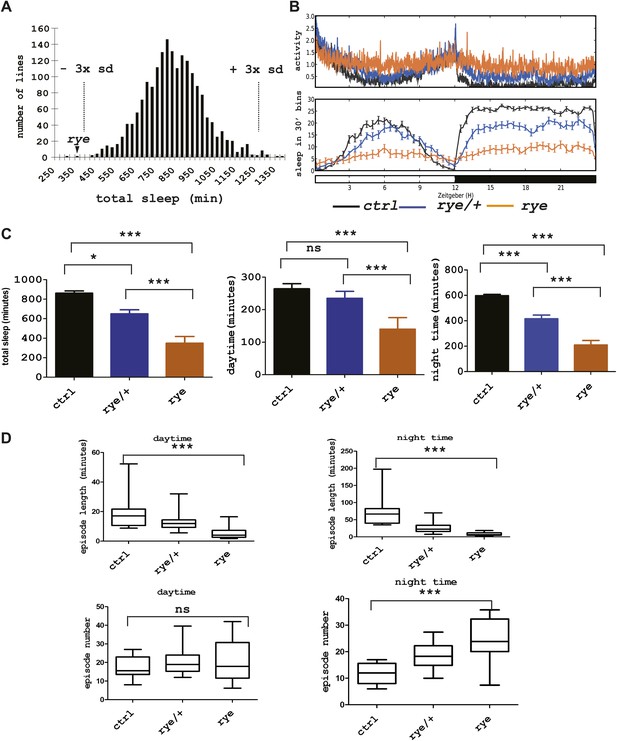
Identification of a short sleep mutant, redeye (rye), through a chemical mutagenesis (EMS) screen.
(A) Histogram depicting average sleep levels in females from homozygous EMS-mutagenized lines (n = 1857). The mean sleep value for each mutant was calculated by assaying sleep in 4–8 individual flies in the presence of 12 hr L-12 hr D cycles. Average sleep for all lines is indicated on the X axis in increments of 25 min. The Y axis depicts the number of mutant lines within each group. The dashed lines mark the sleep values that correspond to plus/minus 3× standard deviation of the mean. The arrowhead indicates the redeye mutant. (B) Top: average activity pattern of females from control lines and from rye mutants recorded in 12hr L-D cycles (n = 14–16 in 5 days). Bottom: sleep profiles with standard error bars. Black: lines isogenized on the 3rd chromosome for chemical mutagenesis; Blue: flies heterozygous for rye and the isogenized control chromosome; Orange: rye homozygotes. (C) Mean values of total sleep, daytime sleep and night-time sleep for rye mutants (Figure 1—source data 1). For each genotype 14–16 flies were assayed over a 5 day period. Bars represent standard error. One-way ANOVA was performed followed by Tukey post hoc analysis. * represents p<0.05, **p<0.01 and ***p<0.001. (D) Sleep quality of rye mutants: Daytime and night-time sleep episode length and episode number are plotted in the box-and-whisker diagram (Figure 1—source data 1). The middle line represents the median value; Bottom and top line of each box represent 25% and 75% respectively; Bottom bar and top bar represent 5% and 95% respectively. ns: not significant, ***p<0.001.
-
Figure 1—Source data 1
Measurement of sleep duration for all EMS mutants and sleep analysis of rye mutants.
- https://doi.org/10.7554/eLife.01473.004
Normal circadian rhythms in rye homozygotes
As the circadian clock regulates the timing of sleep and some clock mutants show reduced sleep (Hendricks et al., 2003), we also assayed rye flies for defects in circadian rhythms. These endogenously generated rhythms are best monitored under constant conditions, in the absence of cyclic environmental cues, so we monitored behavior in constant darkness (DD). rye mutants display robust rest:activity rhythms in DD (∼62% rhythmicity) despite showing prolonged duration of activity (Figure 2A; Table 1). Of the ‘arrhythmic’ rye homozygotes detected, half (6/11) actually displayed a rhythm of ∼12 hr period length, which likely resulted from persistence of bimodal behavior (typically seen in light:dark) in DD. Thus, most rye homozygotes are rhythmic. Consistent with the intact circadian behavior, circadian cycling of the clock protein, PERIOD (PER), is normal in central clock cells of rye mutants. The peaks and troughs of expression, as well as the subcellular localization, at different times of day are similar to those in iso controls (Figure 2B). Thus, PER is largely nuclear at ZT20, entirely nuclear at ZT2, expressed at low levels at ZT8 and undetectable at ZT14. We conclude that only the homeostatic regulation of sleep is affected in rye mutants.
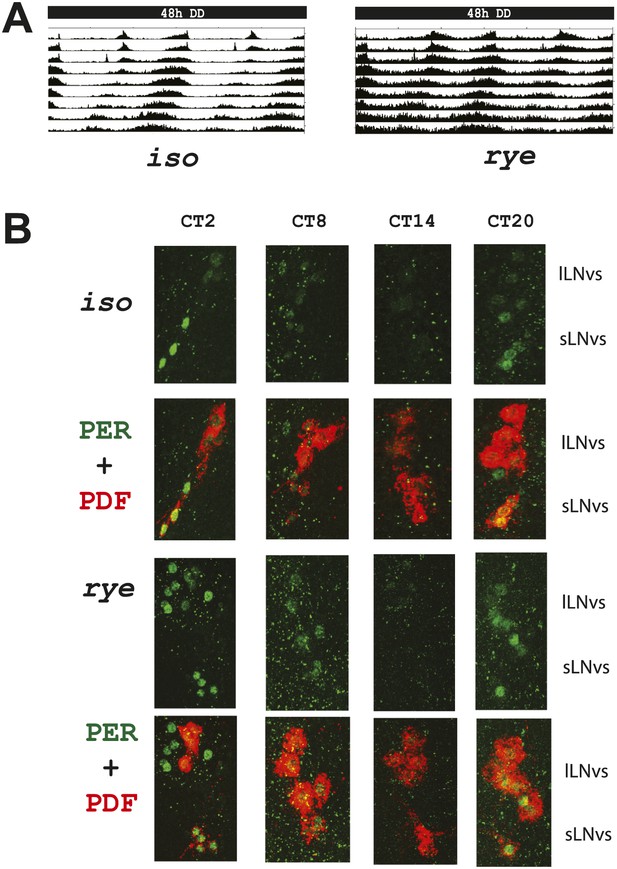
rye homozygotes display normal circadian rhythms and clock protein cycling.
(A) Double plotted activity records of iso controls and rye homozygotes in DD. Rhythmic, but prolonged cross-beam activity was recorded in rye homozygotes. (B) Immunostaining of PERIOD in ventral lateral neurons (LNvs) of iso controls and rye homozygotes at CT time points on the second day of DD following LD entrainment. PER oscillates in these clock neurons in iso as well as in rye homozygotes. PDF labels the small and large LNvs.
Rhythmic behavior of rye homozygotes in constant darkness
| Genotype | Rhythmicity% (n)* | Period ± SEM (h)† | FFT ± SEM† |
|---|---|---|---|
| W1118 (iso) | 93.8% (30/32) | 23.21 ± 0.04 | 0.051 ± 0.005 |
| rye | 62.1% (18/29) | 23.66 ± 0.09 | 0.054 ± 0.005 |
-
*
Flies were entrained to a light–dark cycle (12 hr/12 hr) for 3 days before being moved into constant darkness (DD). Behavior was analyzed from day 3 to day 11 in DD, and flies with a fast fourier transform value (FFT) above 0.01 were considered rhythmic.
-
†
Period lengths and FFT values of rhythmic flies are listed as average value plus minus the standard error of mean (SEM).
Identification of rye as an α subunit of the nicotinic acetylcholine receptor
We identified the sleep-altering lesion in rye mutants through a combination of recombination mapping and deep-sequencing. Despite starting with a newly isogenized line, deep-sequencing identified many nucleotide changes in the rye background relative to the parental control and so considerable mapping was required to pinpoint the relevant mutation. Meiotic recombination analysis positioned rye between two genetic markers, thread and curled, in the centromeric region of the 3rd chromosome. Further mapping studies, using self identified single nucleotide polymorphism (SNP) markers, narrowed it down to ∼11 mega-bases (Figure 3A, Figure 3—source data 1). For finer localization, we utilized the genome-wide sequence data, which provided 40× and 10× genomes worth of nucleotide sequence, corresponding to 95% and 88% coverage, for the rye and iso31 genomes respectively. Following alignment with the published Drosophila genome, unique nucleotide polymorphisms present in rye mutants were pooled together as potential EMS-induced mutations (n = 26,224). Within the 11 Mb region identified through recombination mapping, there were 1457 potential mutations, but only nine that produced an amino-acid change in coding regions. Sanger sequencing allowed us to exclude seven of these. Two turned out to be deep-sequencing errors and five polymorphisms were also present in the wild type iso stock but not detected due to the low coverage of deep-sequencing in this stock. One polymorphism, in a gene annotated CG7320, was at a site that was also polymorphic in iso, although the amino acid change was different in the two strains. Because of the change in iso, which does not produce a sleep phenotype, we considered CG7320 a less likely candidate for rye. The last of the nine changes identified by deep-sequencing was a C-T transition (typical for EMS-induced mutations) that leads to a threonine to methionine change in CG12414 and is present only in rye mutants (Figure 3B). CG12414 encodes an α subunit of a nicotinic acetylcholine receptor (nAchR) (Lansdell and Millar, 2000). Five alternatively spliced variants produced by this gene are predicted to generate three possible open reading frames with one (pC/pG) containing an intact ligand bind domain (LBD) (Figure 3C). The predicted protein is homologous to several nAChR subunits, including α3, α2, α6, α4 and α7 in order of similarity. Alignment of CG12414 with α3 and α7 using the ClustalW2 algorithm shows that the rye mutation is at the junction between the LBD and the trans-membrane domain (TM) (Figure 3D,E). Interestingly, this junction region is highly conserved across species and the specific threonine mutated in rye is conserved in all α7 subunits (Figure 3D).
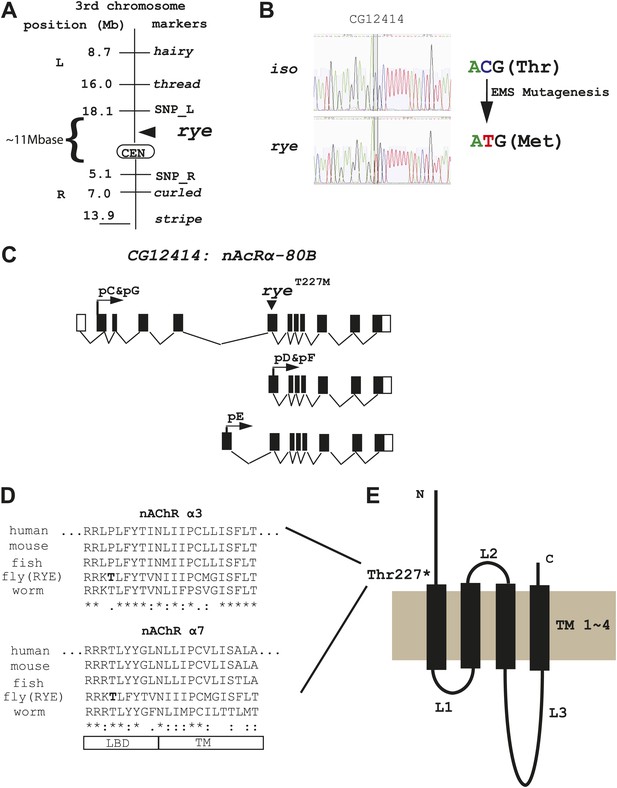
Genetic mapping and genome-wide deep-sequencing reveal a missense mutation in a nicotinic acetylcholine receptor α subunit gene in rye mutants.
(A) Through genetic mapping, using classical phenotypic markers and newly identified SNP markers, rye was mapped between two SNP markers (SNP_L and SNP_R) near the centromere on the 3rd chromosome. (B) Paired-end genomic deep-sequencing of rye mutants identified a missense mutation in CG12414. We confirmed this through Sanger sequencing. This C-T/G-A transition that generated the ryeT227M allele is typical of EMS-induced mutations. (C) Schematic representation of the rye candidate gene, a nicotinic acetylcholine receptor (CG12414: nAcRα-80B). The gene spans ∼100 kb with alternative splicing predicted to produce five transcripts (RC-RG). The spliced forms are predicted to translate into three proteins isoforms (pC-pG). pC(pG) produces the largest protein with the longest ligand binding domain (LBD). (D) Alignment of the partial sequence of Drosophila RYE with nAChR α3 and α7 subunits in other animals using ClustalW2. The region shown is at the boundary of the ligand binding domain (LBD) and transmembrane domain (TM), and is evolutionarily conserved. T227 in RYE is marked in bold form. ‘*’ identity; ‘:’ high similarity; ‘.’ similarity. (E) Protein sequence analysis predicts four transmembrane domains (TM) in RYE, which is typical of most nAChR proteins. RYE appears to contain a single ligand binding domain (LBD) in the extracellular region, and four TMs with three loop regions (L1-L3). The mutated threonine227 is in the LBD, close to the beginning of the TM.
-
Figure 3—Source data 1
Recombination mapping of the rye mutation using a chromosome marked with visible markers h,th,cu,sr,e, and using SNP markers.
- https://doi.org/10.7554/eLife.01473.008
Functional analysis of rye
The genetic mapping and deep-sequencing data combined implicated CG12414 as a candidate gene responsible for the rye sleep phenotype. To verify this function for CG12414, we attempted to rescue rye mutants with a CG12414 transgene. To drive expression in areas that normally express this gene, we cloned the CG12414 promoter region (1.8 kb) and generated a GAL4 construct. This GAL4 is expressed broadly in the Drosophila brain (Figure 4A). We also cloned the CG12414 cDNA into a UAS construct and crossed the GAL4 and UAS transgenes into an iso31 stock to ensure a homogenous genetic background. These transgenes were then introduced into rye mutants. While either transgene alone did not alter the rye short sleep phenotype, the rye promoter driving RYE expression (ryeP > uas-rye) partially rescued the mutant phenotype and restored total sleep to levels seen in rye heterozygotes (Figure 4B). This is expected, given that ryeT227M is partially dominant over wild type. In a wild type background, ryeP > uas-rye does not increase sleep length (data not shown), suggesting that the increase in sleep is specific to the mutant. Thus, we identified the gene responsible for the short sleep phenotype in rye mutants, and henceforth refer to this gene CG12414, previously called nAChR-80B, as redeye.
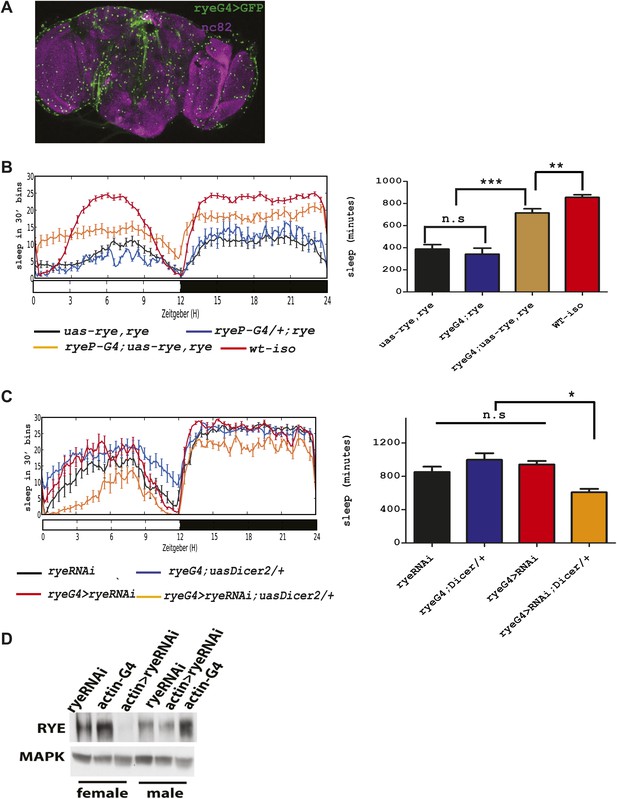
An alpha subunit of the nicotinic acetylcholine receptor accounts for the rye mutant phenotype.
(A) Expression pattern of ryeP-GAL4. Rye-GAL4 was used to express nGFP, which is visualized with an anti-GFP antibody. The anti-nc82 staining marks the neuropil. (B) Expression of the alpha subunit, as described in Figure 3, increases sleep duration in rye mutants (Figure 4—source data 1). Left: sleep profiles, with standard error, of rye mutants and mutants expressing a UAS construct of the putative rye cDNA under control of its own promoter (ryeP-Gal4) in a 12:12 LD cycle. Right: quantification of sleep length. **p<0.01, ***p<0.001. (C) Reduction of rye expression in rye neurons through the expression of an RNAi construct, together with Dicer2, diminishes sleep length (Figure 4—source data 1). Left: sleep profile in a 12:12 LD cycle. Right: quantification of sleep length. *p<0.05. (D) Western blot analysis shows reduced expression of RYE when actin-GAL4 is used to drive rye RNAi (VDRC#11392) with Uas-Dicer2 in female and male flies.
-
Figure 4—Source data 1
Sleep behaviour of transgenically rescued rye mutants and rye RNAi lines.
- https://doi.org/10.7554/eLife.01473.010
To further characterize rye function, we knocked down RYE expression through RNA interference. A rye RNAi line (#11392) from the VDRC stock center in Vienna was crossed into the ryeP-GAL4 line along with a UAS-Dicer2 transgene, included to improve efficacy of the RNAi. Levels of RYE were reduced (Figure 4D), as was the total sleep length (Figure 4C). Since ryeT227M reduces sleep as well (Figure 1), these data suggest that the T227M mutant allele causes loss of RYE function. The sleep phenotype produced by rye knock-down is rather mild in comparison with the phenotype of the homozygous rye mutant, possibly due to inefficient knockdown in relevant cells or because ryeT227M is a dominant-negative mutation. The nicotinic acetylcholine receptor forms a pentameric structure, consisting of five homo-oligomeric or hetero-oligomeric subunits (Miwa et al., 2011), so a dominant negative mutation in any one subunit may well interfere with activity of the complex.
RYE interacts with SSS
We next addressed if rye interacts with other known sleep mutants, in particular with a mutant we identified previously, sleepless (sss), that has an extreme short-sleeping phenotype (Koh et al., 2008). While transheterozygotes of rye and sss (sss/+; rye/+) showed sleep duration comparable to that of rye/+ alone (data not shown), rye heterozygotes carrying a genomic sss transgene, which increases SSS expression above that of wild type controls, showed a further reduction of sleep (Figure 5A). The effect was observed with three independent insertions of the sss transgene, indicating that it was not due to insertion site effects (Figure 5B). We note that this sss transgene does not have a significant effect on sleep in wild type flies although it completely rescues the sleep phenotype of sss mutants (Koh et al., 2008). Thus, the effect is specific to rye mutants.
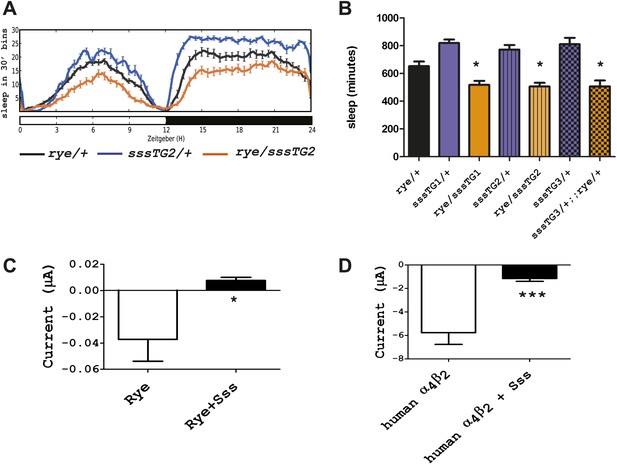
Overexpression of sss exacerbates the rye phenotype by repressing RYE activity.
(A) An extra copy of SSS reduces sleep in rye heterozygotes. Sleep profile of rye heterozygotes with or without a genomic sss transgenic line inserted on the third chromosome (sssTG2) and of sss transgenics alone. (B) Quantification of sleep in the genotypes as indicated (Figure 5—source data 1). TG1 and TG3 are additional independent genomic sss transgenes inserted on the 3rd and X chromosomes respectively. Sleep reduction was observed in rye transheterozygotes with all three sss transgenic lines. *p<0.05. (C) Heterologous expression of SSS reduces current following ACh application in Xenopus oocytes expressing Drosophila RYE and human nAChR β2 (Figure 5—source data 1). *p<0.05. (D) Heterologous expression of SSS reduces current following ACh application in Xenopus oocytes expressing human nAChR α4β2 (Figure 5—source data 1). ***p<0.001.
-
Figure 5—Source data 1
Sleep analysis of rye mutants overexpressing sss, and whole-cell membrane current recording of Xenopus oocytes.
- https://doi.org/10.7554/eLife.01473.012
SSS is a GPI-anchored protein that regulates the Shaker potassium channel and its predicted structure resembles that of a specific class of toxins (Wu et al., 2010; Dean et al., 2011). It is most similar to the mammalian Lynx-1 protein, which is known to inhibit activity of the nicotinic acetylcholine receptor (Miwa et al., 2011). The fact that SSS exacerbated the phenotype of flies heterozygous for rye, which have reduced activity of nAChR, suggested that SSS also regulates nAChRs. We tested a possible Lynx-1-like function of SSS in regulating nAChR activity by using a heterologous expression system, specifically Xenopus laveis oocytes. For functional expression of wild type RYE, we expressed it with a human β2 subunit, as insect β subunits typically do not work in heterologous systems (Millar and Lansdell, 2010). cRNAs encoding sss, wild type rye and human β2 were injected into oocytes, and 4–7 days later whole cell currents were measured following application of ACh (Kuryatov and Lindstrom, 2011). Heterologous expression of insect nAChRs has, in general, been challenging (Millar, 2009), which accounts for the low current values observed (Figure 5C). However, we still detected a significant reduction of nAChR current upon co-expression with SSS (Figure 5C). We also co-expressed in vitro transcribed sss and human α4 and β2 subunits and found that SSS also inhibits the robust current produced by the human receptor complex (Figure 5D). Thus, in addition to its sleep-promoting role reported previously, SSS may also promote wake by repressing activity of nAChRs. As discussed below, loss of the sleep-promoting role has a dominant effect in sss mutants.
RYE is expressed cyclically in association with the sleep state
We next asked if levels of rye vary over the course of the day. Groups of wild type flies were housed in a 12 hr-light and 12 hr-dark incubator and harvested at different Zeitgeber times (ZT). Zeitgeber time defines time based on the environmental stimulus, so ZT0 corresponds to light-on and ZT12 to light-off. RNA was extracted and subjected to quantitative PCR analyses. While period (per), a well-known circadian clock component, oscillates with a circadian rhythm (Zheng and Sehgal, 2012), rye mRNA levels were found to be constant (Figure 6A).
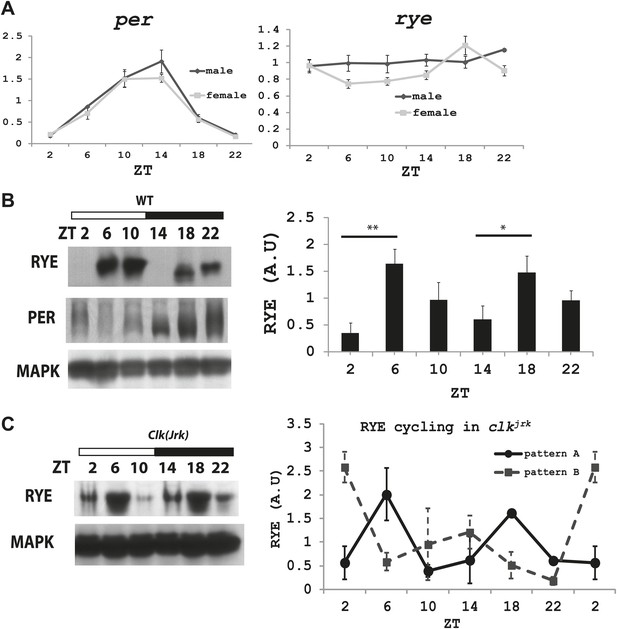
The RYE protein is expressed cyclically in association with the sleep state, but independently of the circadian clock.
(A) Quantitative PCR analyses show oscillations of per mRNA (left panel), but constant levels of rye mRNA (right panel). actin was used as a normalization control (Figure 6—source data 1). (B) Left: a representative western blot of head extracts from wild type flies shows cyclic expression of RYE with two daily peaks, in the middle of the day (ZT6-10) and in the middle of the night (ZT18-22), corresponding to the sleep state (Figure 1B). PER, in contrast, shows only one daily peak. MAPK was used to control for loading. Right: densitometry quantification of western blots with error bars representing standard error (n = 8). RYE value at ZT0 is set as 1 (Figure 6—source data 1). *p<0.05, **p<0.01. (C) RYE cycling is similar to wild type in Clkjrk flies, indicating that cycling per se does not require a functional clock. However, the phase is variable, so for quantification purposes five independent experiments were split into two groups of roughly similar phase (Figure 6—source data 1).
-
Figure 6—Source data 1
qPCR analysis of per and rye expression during LD cycle and densitometry quantification of RYE expression.
- https://doi.org/10.7554/eLife.01473.014
To examine protein levels of RYE, we generated an antibody against the cytoplasmic L3 domain of RYE (Figure 3E), which is the most variable region across all nAChR subunits (Lindstrom, 2003). On western blots, the antibody recognizes a band (or sometimes a doublet) of ∼60 Kd, which is the size predicted for the full-length isoform. Surprisingly, in contrast to the flat mRNA levels, levels of RYE protein show a robust rhythm with two peaks each day, one at around ZT6-10 and the other at ZT18-22 (Figure 6B, Figure 6—figure supplement 1). Interestingly, these correspond to the two daily times of sleep, the afternoon siesta and night-time sleep (Figure 1B), suggesting that RYE is expressed in phase with sleep. The cycling of RYE persists in constant darkness, as do rhythms of sleep (data not shown). However, RYE does not appear to be regulated like other cycling clock components, such as PER, which usually show only one peak (Figure 6B). Furthermore, we found that RYE continues to cycle in Clkjrk flies that lack a circadian clock, although the phase of RYE expression tends to be more variable in these flies (Figure 6C, Figure 6—figure supplement 2). These data suggest that the RYE oscillation per se does not require process C of the two process model, but may depend upon process S (sleep homeostasis). However, the circadian clock may function to provide more precise timing of RYE expression, perhaps through its control of sleep onset and offset.
Expression of RYE is under control of the sleep homeostat
Increases in RYE expression could occur as a consequence of sleep, and thereby reflect the sleep state, but in that case levels should remain high throughout the sleep state. Given that RYE levels do not remain high throughout the night or through the afternoon siesta, another possibility is that elevations in RYE correspond to sleep drive and so occur at the time of sleep onset. To test this idea, we assayed RYE expression in short-sleeping mutants. These mutants have low sleep levels, but probably have high sleep drive that cannot be implemented. As shown in Figure 7, for the most part RYE expression exhibits two peaks in these mutants, but the phase is more variable (Figure 7—figure supplements 1–3), indicating that defects in sleep behavior affect the RYE expression pattern. Interestingly, overall RYE values are higher in fumin (Kume et al., 2005; Ueno et al., 2012), sss (Koh et al., 2008) and insomniac (Stavropoulos and Young, 2011) mutants than in wild type controls. As noted above, the sss mutation affects channel activity, fumin is a mutation in the dopamine transporter (short sleep is thought to result from the high levels of dopamine in the synaptic cleft) and insomniac is thought to affect protein turnover. The one feature these mutants have in common is short sleep, yet they all have elevated RYE expression during an LD cycle (Figure 7A–C). Based upon these data, we suggest that RYE reflects sleep need. It is elevated at the time of sleep onset and is further increased in short-sleeping mutants because they have high sleep need.
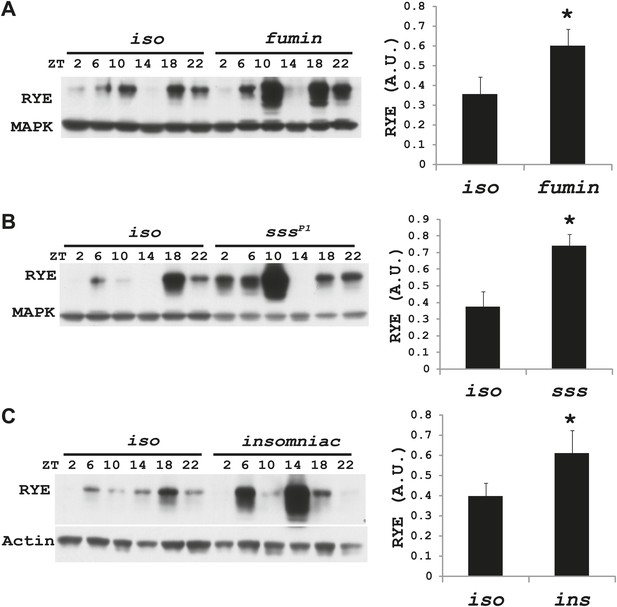
RYE levels are elevated in short-sleeping mutants.
(A) Western analyses of RYE expression over the course of a day in iso controls and fumin mutants. While RYE still cycles in fumin, the mean value of all six daily time points from three independent experiments (n = 3) shows that overall levels of RYE are higher than in wild type controls (Figure 7—source data 1). *p<0.05. (B) Western analyses of RYE expression over the course of a day in iso controls and sss mutants. As in A, the quantification reflects the mean across the day from three independent experiments (n = 3). *p<0.05. (C) Western analyses of RYE expression over the course of a day in iso controls and insomniac mutants. Quantification as above (n = 3). *p<0.05.
-
Figure 7—Source data 1
Densitometry quantification of RYE expression in short sleep mutants.
- https://doi.org/10.7554/eLife.01473.018
If the expression of RYE changes in response to the accumulation of sleep need, it should also be affected by acute sleep deprivation. We mechanically deprived flies of sleep in the second half night (6 hr) in an LD cycle, and confirmed, through the presence of a substantial sleep rebound the following morning, that the flies were successfully deprived (Figure 8A; Koh et al., 2008). RYE levels were assayed during the 5 hr sleep rebound window. In the non-deprived control, RYE levels were low at ZT0 and high at ZT5, as noted above. Sleep-deprived flies had high levels of RYE immediately following deprivation (ZT0), but levels were low at ZT5 (Figure 8A, Figure 8—figure supplement 1). This fits the profile for sleep drive, which is expected to be high at the end of deprivation, but then dissipated over the course of rebound sleep.
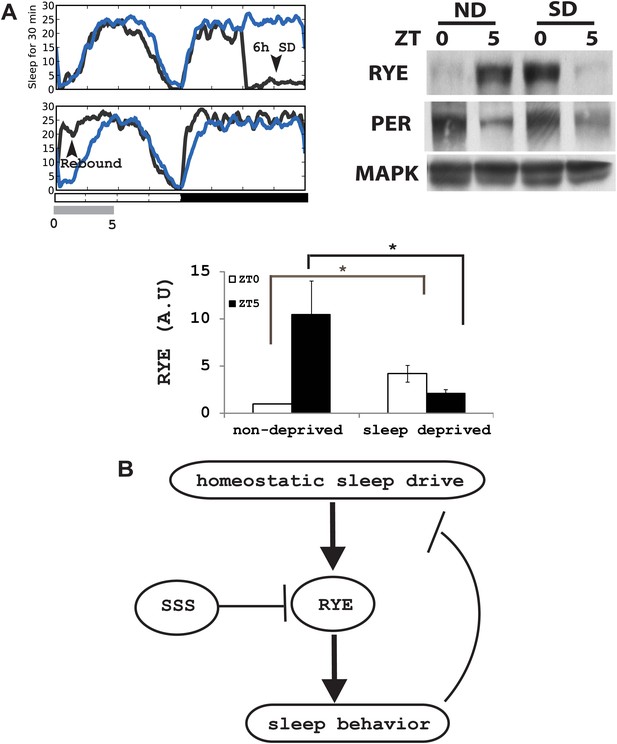
RYE expression is under control of the sleep homeostat.
(A) Left: sleep profile of sleep deprived flies. Blue line: non-deprived controls, Grey line: sleep deprived flies. Arrowheads point to the 6 hr sleep deprivation (SD) window (ZT18-24) and sleep rebound the following morning (ZT0-5). Right: a representative western blot of fly head samples at ZT0 and ZT5 in sleep-deprived (SD) animals and non-deprived (ND) controls. RYE levels are high immediately following sleep deprivation (SD0) and dissipate after sleep rebound (SD5). In contrast, the circadian clock is not affected by SD, since PER cycling is comparable in the SD group and the non-deprived control (ND). Densitometry quantification of the western data (n = 5) is shown below with error bars showing standard error (Figure 8—source data 1). *p<0.05. (B) Model for the role of RYE in the homeostatic regulation of sleep: We propose that RYE is required to maintain sleep, and posttranslational regulation of RYE reflects homeostatic sleep drive. Sleep need builds up during wakefulness and upregulates levels of RYE, which are essential to maintain sleep behavior. Homeostatic drive dissipates during sleep, and levels of RYE are reduced, leading to wakefulness. SSS represses RYE activity, thus acting as a wake-promoting factor in this particular context.
-
Figure 8—Source data 1
Densitometry quantification of RYE expression after SD.
- https://doi.org/10.7554/eLife.01473.023
Discussion
The molecular mechanism of sleep homeostasis is a mystery and a subject of intense research in the sleep field (Suzuki et al., 2013). In addition to the investigation of mechanisms underlying sleep drive, considerable effort is being put into identifying biomarkers of sleep need. Based on what we know about the so-called sleep homeostat, which consists of increasing sleep pressure during wakefulness and dissipation of such pressure following sleep, we suggest that a component or direct output of the homeostat should satisfy three criteria: (i) the gene product should regulate the sleep:wake cycle (i.e., genetic alleles of this gene should have some sleep phenotype); (ii) expression levels or activity of the gene product should go up during wakefulness or during sleep deprivation and (iii) expression levels or activity should decrease after sleep. The function of RYE and the molecular kinetics of the RYE protein largely satisfy these criteria. However, while RYE builds up during sleep deprivation, it does not accumulate gradually over the wake period in a daily cycle. Rather, it displays a marked increase close to the time of sleep onset, suggesting that it is not a central component of the homeostat, but responds to an upstream homeostatic signal, perhaps when that signal reaches a certain threshold. The fact that over-expression of RYE does not promote sleep also supports the idea that it is not the sleep-inducing homeostatic signal (further discussed below). Nevertheless, RYE is not simply a sleep output gene or sleep biomarker. It is required for implementation of signals from the homeostat and it functions to maintain sleep. Thus, we propose that rye is a sleep-regulating gene immediately downstream of the homeostat.
RYE expression reflects sleep need
We suggest that RYE represents a molecular correlate of delta power, a characteristic of an electroencephalogram (EEG) that reflects sleep drive. Recently, a few other molecules were reported to change with sleep drive, but the effects were at the level of the mRNA, the magnitude of the increase was less than we report here for RYE and loss of the molecules did not affect baseline sleep duration (Seugnet et al., 2006; Maret et al., 2007; Naidoo et al., 2007). In addition, only one is expressed cyclically (Nelson et al., 2004), indicating that others reflect sleep drive only under pathological conditions of sleep deprivation. RYE levels oscillate robustly in a daily cycle, although the phase is not as coherent as seen for circadian clock proteins. The timing of the peak varies within a temporal range, such that there is almost always a daytime peak and a night-time peak but not necessarily at the exact same time (Figure 6—figure supplement 1). We suggest that RYE cycles under control of the sleep homeostat, which may not time behavior as precisely as the circadian clock, perhaps because sleep can be influenced by many factors. The variability in RYE cycling is particularly pronounced in short-sleeping mutants and in the ClkJrk circadian clock mutant (Figure 6—figure supplement 2, Figure 7—figure supplements 1–3), suggesting that the clock does influence RYE expression although it is not required for its cycling in an LD cycle. Interestingly, RYE cycles exclusively at the level of the protein, indicating translational or post-translational mechanisms. It is worth noting that a recently identified sleep regulator, Insomniac, is a component of specific protein degradation pathways in the cell (Stavropoulos and Young, 2011). Although our study indicates that RYE cycling does not require Insomniac (Figure 7C), it is possible that it is regulated by other protein turn-over machinery. Thus, translational/posttranslational regulation appears to be part of the mechanism of sleep homeostasis.
RYE is required to maintain the sleep state
We show that RYE not only reflects sleep drive, but is also required for sleep maintenance (Figures 1, 4 and 8). Given that RYE is induced by sleep deprivation and it promotes sleep, one might expect over-expression of the protein to increase sleep. However, transgenic expression of rye in a wild type background does not increase sleep, suggesting that while rye is necessary, it is not sufficient for sleep onset. We cannot exclude the possibility that RYE functions together with other signals as part of the sleep-inducing homeostatic drive. On the other hand, it is also possible that transgenic expression does not produce adequate amounts of RYE protein in relevant cells. This might be the case if RYE is tightly regulated at the level of protein stability. For the moment, though, we prefer the parsimonious explanation noted above, that RYE is not part of the homeostat, but immediately downstream of it.
Acetylcholine signaling has been long proposed as an arousal factor, as the nAChR complex is a cation channel that normally promotes neuronal activity and ACh is released during wakefulness in mammals (Platt and Riedel, 2011). In contrast, our study indicates that at least one nAChR subunit (RYE) promotes sleep in the fly. There are more than 10 paralogs of nAChR subunits in the fly genome. One possibility is that RYE is expressed specifically in sleep promoting neurons, while other subunits of AChRs are in wake-promoting cells. An increase in ACh during wakefulness may contribute to the accumulation of sleep drive and to the increase in RYE (Lindstrom, 2003). Alternatively, sleep drive may increase RYE independently of ACh, but in either scenario, RYE then promotes sleep. The precise site of RYE action is currently not known. rye-gal4 driven GFP is expressed widely in the brain (Figure 4A), but we cannot be sure that endogenous RYE is as widespread, as our antibody was not effective in immunohistochemistry experiments, and we find that GAL4 drivers are often quite promiscuous (unpublished observations).
SSS inhibits RYE activity and promotes arousal in a sensitized background
SSS was previously identified as a sleep promoting factor, essential for maintaining baseline sleep and for homeostatic rebound (Koh et al., 2008). An interaction between rye and sss is therefore not surprising. What is surprising is that overexpression of SSS promotes wakefulness in ryeT227M heterozygotes (Figure 5A,B). SSS is a GPI-anchored protein that functions as a neuronal modulator. Previous studies indicate that SSS promotes activity of the voltage-gated potassium channel, Shaker (Wu et al., 2010). In this study, we report that SSS acts like a brake on nAChR (RYE) activity (Figure 5C,D), as does Lynx-1, a SSS-like molecule, in mammals (Miwa et al., 2011). Although the data we show for Drosophila receptors used only the RYE α subunit, it is likely that SSS also inhibits activity of other Drosophila nAChR receptors. As both sssP1 (a null mutation) and sssP2 (a hypomorphic allele) are short-sleeping mutants (Koh et al., 2008), we propose that the overall effect of SSS is to promote sleep. The reduced sleep in sss mutants probably results from an increase of neuronal excitability, through inactivation of potassium channels (Shaker) (Dean et al., 2011), or from hyperactivity of nAChR channels in wake-promoting neurons. Thus, typically the sleep-inhibiting effect of SSS, mediated through RYE, is masked by these other more dominant influences. However, in a sensitized background (i.e., rye/+), this effect is evident. RYE promotes sleep, and so loss of RYE results in a decrease in sleep, which is further impacted by SSS overexpression (Figure 8B).
We note that there are some caveats to these data. For instance, the ryeT227M allele could confer a neomorphic function that accounts for the interaction with sss. Likewise, the effects in oocytes could be non-physiological, not necessarily reflecting what happens in the fly brain. However, given that we observe interactions in these two very different types of assays, and both assays indicate repression of nAchR function by SSS, which is the effect predicted from the role of the mammalian SSS-like protein, Lynx1, we believe SSS does indeed regulate nAchRs such as RYE. Interactions between SSS and nicotinic acetylcholine receptors are also reported by recent work from another laboratory (W Joiner, personal communication).
It is interesting that genes identified through independent genetic screens in Drosophila are turning out to interact with one another. SSS and Shaker were isolated independently as sleep-regulating genes (Cirelli et al., 2005; Koh et al., 2008), and subsequently shown to interact, and now we find that RYE interacts with SSS. Given that each of these genes represents a relatively infrequent hit in an unbiased screen, the interactions suggest that genetic approaches are converging upon specific sleep-regulating pathways. Interestingly, a recent GWAS study for sleep-altering loci in humans identified significant effects of SNPs in an nAchR subunit as well as in a regulatory subunit of Shaker, suggesting that these mechanisms are also conserved across species (Allebrandt et al., 2013).
Materials and methods
Generation of an isogenic 3rd chromosome stock
Request a detailed protocolWild type iso31 (Ryder et al., 2004) was crossed with TM2/TM6C, Sb (Bloomington stock #5906), and a single male progeny was selected and crossed with #5906 to balance the 3rd chromosome and generate a line isogenic for this chromosome.
Behavioral analysis
Request a detailed protocolFlies were housed in Percival incubators (Perry, IA) and beam–break activity was recorded with the Trikinetics DAM system (http://www.trikinetics.com/). Pysolo (Gilestro and Cirelli, 2009) software was used to analyze and plot sleep patterns. Sleep deprivation was achieved through repeated mechanical shaking (2 s randomized shaking in every 20-s interval).
Mapping analysis
Request a detailed protocolA 3rd chromosome marker line h, th, cu, sr, e (Bloomington stock #576) was crossed to rye/TM6C, Sb, and heterozygote female progeny (h, th, cu, sr, e/rye) were further crossed to TM2/TM6C, Sb (#5906). A single recombinant male offspring was back-crossed to #576 to score the genotype of recessive markers and also back-crossed to rye for behavioral analysis to score the rye genotype. Overlap in the genotypes of recombinants narrowed down the location of the rye mutation to the region between markers th and cu.
Genomic DNA of homozygous recombinants was subject to SNP analysis. The primer set 5′TGTTTAGTGGTGTTGTGTGAGC3′ and 5′GCCGAGTGTCATCGCCTTTG3′ for SNP_L and the primer set 5′AAAGGTCATCTTGCTTCGGAGTTG3′ and 5′GGAGTGGCTTCCTCGTCATC3′ for SNP_R were used for PCR amplification. Nucleotide polymorphisms were identified between iso and #576. Thus, those recombinants were scored accordingly and rye was further narrowed down to a region between those two SNP markers.
Sequencing and cloning
Request a detailed protocolFragmented genomic DNA (∼400 bp in size) obtained through the Covaris instrument (Woburn, MA) was subjected to Illumina paired-end DNA library preparation. The libraries were amplified for 10 PCR cycles prior to Illumina Hi-Seq analyses. SNP calling analyses identified nucleotide polymorphisms in both rye mutants and iso flies.
The primer set 5′GGCTCGATGTGCTTTCAAGAGTTC3′ and 5′CATGCCAGATGAGTGCGTTTC3′ was used to clone the rye promoter region from genomic DNA derived from the iso stock. The primer set 5′TTTAGGCTTAGTCCGCTACC 3′ and 5′AATGTCGTGGTTTGAAGTGC 3′ was used to clone the full length rye cDNA. The PhiC31 integration system was used for injection. The rye promoter construct was specifically inserted onto the 2nd chromosome and the Uas-rye construct was integrated on the 3rd chromosome.
Generation of RYE antibodies
Request a detailed protocolThe primer set 5′ATGCATCATCACCATCACCATAGTATCTGCGTGACGGTTGTTG3′ and 5′GTGGTGGTGCACAACTGCCAACGTGAATATCC 3′ was used to clone the L3 epitope of RYE. A GST-RYEL3 fusion protein was expressed in Escherichia coli, excised from a PAGE gel and injected into rats for antibody generation.
Extractions and measurements of DNA, RNA and protein
Request a detailed protocolDrosophila genomic DNA: flies (3–15) were frozen and homogenized in DNA extraction buffer. After LiCl/KAc precipitation, supernatants were subject to isopropanol precipitation. DNA pellets were dissolved in TE for sequencing analyses and PCR amplification.
RNA: adult flies were collected at indicated time points and fly heads (10∼20) were subject to Trizol extraction (Ambion, Life Technologies, Grand Island, NY). cDNA libraries were made through high-capacity cDNA reverse transcription kits (Applied Biosystems, Life Technologies, Grand Island, NY). Quantitative PCR analysis was performed using SYBR green reagents in the Applied Biosystems 7000 sequence detection system. The primer set 5′AGTTGAATGGAAGCCACCAGC3′ and 5′TGTTCATCCATGTGCCTCAG3′ was used for qPCR analysis of rye expression.
Protein: flies were collected at indicated time points and fly heads (∼5) were prepared for protein extraction and for Western analyses as previously described (Luo and Sehgal, 2012).
Immunocytochemistry assays
Request a detailed protocolAdult fly brains were dissected in cold PBS buffers at indicated CT time points. Brains were fixed in 4% PFA for 30 m and incubated with 5% Normal Donkey Serum (NDS) for 1 hr at the blocking step. Incubation with primary rabbit anti-PER (1:1000), mouse anti-PDF (1:1000) and rabbit anti-GFP, mouse anti nc82 was performed overnight in the cold room. After extensive washing, brains were incubated with the donkey anti-rabbit or anti-mouse secondary antibody (1:1000) for 2 hr. Images were taken under a Leica confocal microscope.
Electrophysiological current recording in Xenopus oocytes
Request a detailed protocolOocytes were removed surgically from Xenopus laevis and placed in an OR-2 solution containing 82.5 mM NaCl, 2 mM KCl, 1 mM MgCl2, 5 mM HEPES, and pH 7.5. They were defolliculated in this buffer containing 2 mg/ml collagenase type IA (Sigma, St. Louis, MO) for 1.5 hr. After defolliculation, oocytes were incubated at 18°C in 50% L15 (Invitrogen, Carlsbad, CA) 10 mM HEPES, pH 7.5, 10 units/ml penicillin, and 10 g/ml streptomycin at 18°C. 3–6 days after injection, whole-cell membrane currents evoked by acetylcholine were recorded in oocytes at room temperature with a standard two-electrode voltage-clamp amplifier (Oocyte Clamp OC-725; Warner Instrument, Hamden, CT). Recordings were performed at a holding potential of −50 mV. All perfusion solutions contained 0.5M atropine to block responses of endogenous muscarinic AChRs that might be present in oocytes. Acetylcholine was applied by means of a set of 2-mm glass tubes directed to the animal pole of the oocytes. Application was achieved by manual unclamping/clamping of a flexible tube connected to the glass tubes and to reservoirs with the test solutions. The recording chamber was perfused at a flow rate of 15–20 ml/min. cRNAs for human AChR subunits 4 and 2 and Drosophila sss were synthesized in vitro using SP6 RNA polymerase (mMESSAGEmMACHINETM, Ambion). Oocytes were injected with 5 ng of cRNA of each of the subunits.
References
-
The genetic and molecular regulation of sleep: from fruit flies to humansNature Reviews Neuroscience 10:549–560.https://doi.org/10.1038/nrn2683
-
Reduced sleep in Drosophila Shaker mutantsNature 434:1087–1092.https://doi.org/10.1038/nature03486
-
Drosophila QVR/SSS modulates the activation and C-type inactivation kinetics of Shaker K(+) channelsThe Journal of Neuroscience 31:11387–11395.https://doi.org/10.1523/JNEUROSCI.0502-11.2011
-
pySolo: a complete suite for sleep analysis in DrosophilaBioinformatics 25:1466–1467.https://doi.org/10.1093/bioinformatics/btp237btp237
-
Gender dimorphism in the role of cycle (BMAL1) in rest, rest regulation, and longevity in Drosophila melanogasterJournal of Biological Rhythms 18:12–25.https://doi.org/10.1177/0748730402239673
-
Kv3 potassium channels control the duration of different arousal states by distinct stochastic and clock-like mechanismsEuropean Journal of Neuroscience 23:1567–1574.https://doi.org/10.1111/j.1460-9568.2006.04672.x
-
Dopamine is a regulator of arousal in the fruit flyThe Journal of Neuroscience 25:7377–7384.https://doi.org/10.1523/JNEUROSCI.2048-05.2005
-
Nicotinic acetylcholine receptors of muscles and nerves: comparison of their structures, functional roles, and vulnerability to pathologyAnnals of the New York Academy of Sciences 998:41–52.https://doi.org/10.1196/annals.1254.007
-
Homer1a is a core brain molecular correlate of sleep lossProceedings of the National Academy of Sciences of the United States of America 104:20090–20095.https://doi.org/10.1073/pnas.0710131104
-
Characterisation of insect nicotinic acetylcholine receptors by heterologous expressionAdvances in Experimental Medicine and Biology 683:65–73.https://doi.org/10.1007/978-1-4419-6445-8_6
-
A role for the molecular chaperone protein BiP/GRP78 in Drosophila sleep homeostasisSleep 30:557–565.
-
The cholinergic system, EEG and sleepBehavioural Brain Research 221:499–504.https://doi.org/10.1016/j.bbr.2011.01.017
-
Identification of a biomarker for sleep drive in flies and humansProceedings of the National Academy of Sciences of the United States of America 103:19913–19918.https://doi.org/10.1073/pnas.0609463104
-
Behavioral and biochemical dissociation of arousal and homeostatic sleep need influenced by prior wakeful experience in miceProceedings of the National Academy of Sciences of the United States of America 110:10288–10293.https://doi.org/10.1073/pnas.1308295110
-
Identification of a dopamine pathway that regulates sleep and arousal in DrosophilaNature Neuroscience 15:1516–1523.https://doi.org/10.1038/nn.3238
-
Speed control: cogs and gears that drive the circadian clockTrends in Neurosciences 35:574–585.https://doi.org/10.1016/j.tins.2012.05.007
Article and author information
Author details
Funding
Howard Hughes Medical Institute
- Amita Sehgal
The funder had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Acknowledgements
We are grateful to Christine Quake for technical support of this project, Matt Kayser, Dan Cavanaugh, Katarina Moravcevic and Christine Dubowy for critical reading of the manuscript and other members of the laboratory for useful discussions. We thank William Joiner for communicating results prior to publication. We also thank Natania Field for suggesting the name, redeye. AS is an Investigator of the HHMI.
Version history
- Received: September 4, 2013
- Accepted: December 12, 2013
- Version of Record published: February 4, 2014 (version 1)
Copyright
© 2014, Shi et al.
This article is distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use and redistribution provided that the original author and source are credited.
Metrics
-
- 2,476
- views
-
- 407
- downloads
-
- 77
- citations
Views, downloads and citations are aggregated across all versions of this paper published by eLife.
Download links
Downloads (link to download the article as PDF)
Open citations (links to open the citations from this article in various online reference manager services)
Cite this article (links to download the citations from this article in formats compatible with various reference manager tools)
Further reading
-
A protein called RYE – short for Redeye – has a central role in the regulation of sleep.
-
- Cell Biology
- Neuroscience
Alternative RNA splicing is an essential and dynamic process in neuronal differentiation and synapse maturation, and dysregulation of this process has been associated with neurodegenerative diseases. Recent studies have revealed the importance of RNA-binding proteins in the regulation of neuronal splicing programs. However, the molecular mechanisms involved in the control of these splicing regulators are still unclear. Here, we show that KIS, a kinase upregulated in the developmental brain, imposes a genome-wide alteration in exon usage during neuronal differentiation in mice. KIS contains a protein-recognition domain common to spliceosomal components and phosphorylates PTBP2, counteracting the role of this splicing factor in exon exclusion. At the molecular level, phosphorylation of unstructured domains within PTBP2 causes its dissociation from two co-regulators, Matrin3 and hnRNPM, and hinders the RNA-binding capability of the complex. Furthermore, KIS and PTBP2 display strong and opposing functional interactions in synaptic spine emergence and maturation. Taken together, our data uncover a post-translational control of splicing regulators that link transcriptional and alternative exon usage programs in neuronal development.





