Meta-Research: Incidences of problematic cell lines are lower in papers that use RRIDs to identify cell lines
Figures
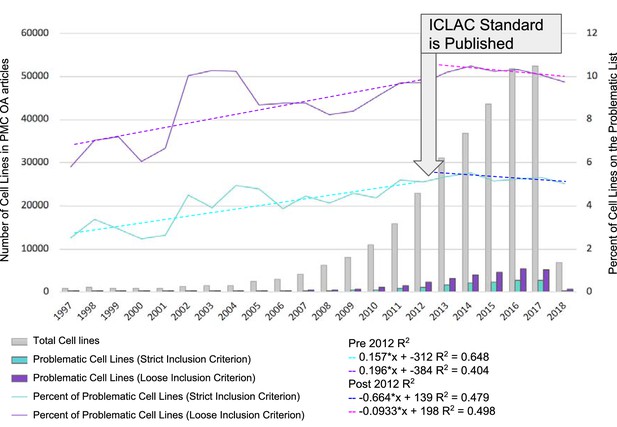
Identification of misidentified cell lines.
The number of cell lines used in PubMed Central articles available for text mining is shown as a function of year. The names of cell lines were matched using two criteria, strict and loose. The strict criterion constitutes an exact match where the name used by the researchers and detected by SciScore is on the list of ICLAC register of misidentified cell lines. The loose criterion was calculated by adding a wild-card character (*) to the end of all names found by SciScore, and matching the names and synonyms on the ICLAC list. The graph is divided into two sections: before and after 2012. 2012 was chosen as the year to break the graph because the publication of the authentication standard and the formation of ICLAC occurred that year (Masters, 2012).
-
Figure 1—source data 1
Underlying data for Figure 1.
- https://doi.org/10.7554/eLife.41676.003
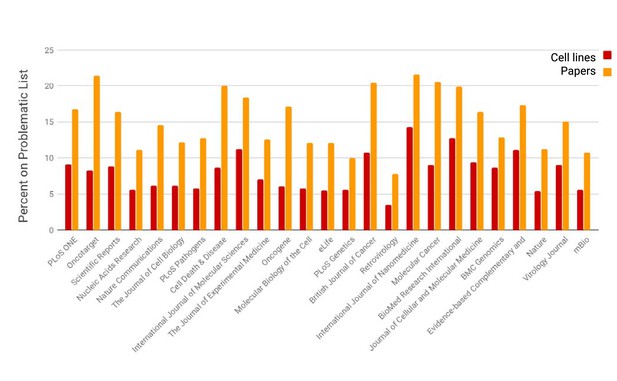
The prevalence of open access papers containing one or more cell lines found on the problematic list.
Journals are sorted from left to right by the number of cell lines detected by SciScore (only the top 25 journals are shown for presentation purposes; data for all journals is given in Figure 2—source data 1). Each bar represents the percent of cell lines (red) or papers (orange) that are on the problematic cell-line list. Cell line presence on the misidentified list is scored by the edit distance metric, which skips all special characters such as spaces and dashes and assumes that any string that contains the same letters and numbers is an edit distance of 0 (e.g., EF 1 = EF-1). Journals that published papers under a license not allowing text mining are not represented here.
-
Figure 2—source data 1
Data on problematic cell lines for all journals.
- https://doi.org/10.7554/eLife.41676.005
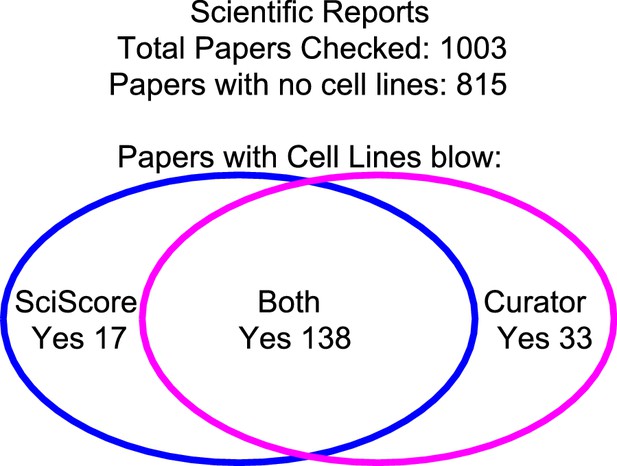
The integrity of SciScore for finding papers with cell lines.
A manual review of 1,003 papers from the journal Scientific Reports showed a 95% agreement between the curator and the SciScore algorithm. Both the curator and SciScore detected a cell line in 138 articles, and no cell line in 822. Of 1,003 papers, 50 represent a disagreement (false positives and false negatives).
-
Figure 3—source data 1
Underlying data for Figure 3.
- https://doi.org/10.7554/eLife.41676.007
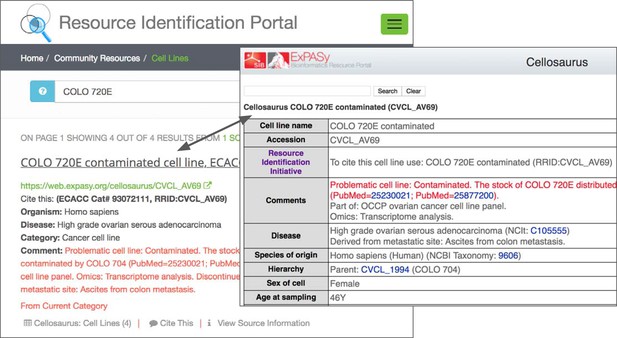
The warning message on the RRID portal and the Cellosaurus database present a misidentified cell line, COLO 720E.
However, this warning does not originate at either the Cellosaurus (the naming resource for problematic cell lines) or the RRID sites; it simply reflects the information available at ICLAC.org. ICLAC members examine publications and test data to reach a conclusion, and then disseminate this on their website via a spreadsheet available to everyone for download. The Cellosaurus database picks up these data, working closely with ICLAC, and updates their entries. The data are then passed to the RRID portal, where it is displayed for researchers searching for cell lines, among other resources. Cellosaurus and the RRID portal strive to make all new data available as quickly as possible.
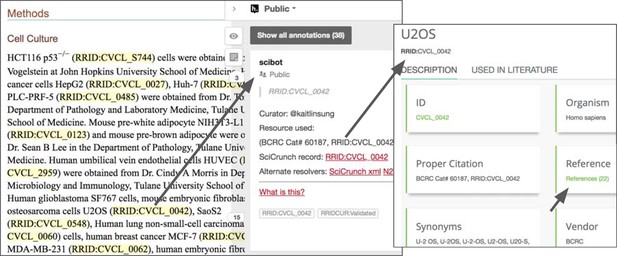
An example of a public annotation using the hypothes.is platform.
Note, all data made in the public channel, such as RRID resolution data, are ported daily to the CrossRef Event database for developers, providing additional ways of making these data FAIR (that is, findable, accessible, interoperable and reusable). Information about this cell line is accessible to readers with one click, including papers that use the cell line and original reference. For journals like eLife, which typeset the RRIDs with live links, hypothesis is not necessary to access the information about cell lines. Based on the paper Liao et al., 2017 using the hypothes.is platform.
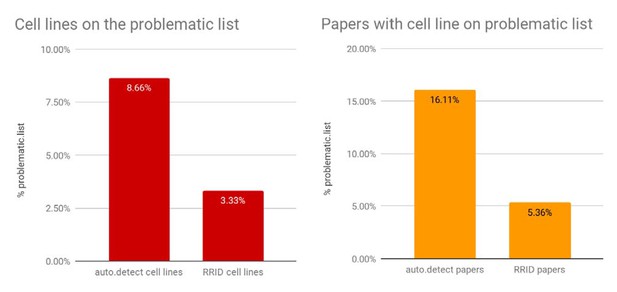
Percentages of papers with cell lines found on the problematic list.
The "auto.detect.cell" lines data come from the edit distance metric, same as Figure 2; n=305,161; the RRID cell lines are based on 1,502 cell lines. The "auto.detect" papers percentage is based on n=150,459 unique papers, where the problematic cell-line list is detected based on the edit distance metric. The RRID papers percentage is based on n=634 papers.
-
Figure 6—source data 1
Data on number of misidentified cell lines per year.
- https://doi.org/10.7554/eLife.41676.011
Additional files
-
Supplementary file 1
List of RRID papers.
- https://doi.org/10.7554/eLife.41676.013
-
Supplementary file 2
List of problematic cell lines extracted from Cellosaurus Version 25 (March 2018).
- https://doi.org/10.7554/eLife.41676.014
-
Supplementary file 3
Curator-SciScore-disagreement - the false negatives (33 papers) found by the curator.
- https://doi.org/10.7554/eLife.41676.015
-
Supplementary file 4
Details for the 50 RRID papers.
- https://doi.org/10.7554/eLife.41676.016
-
Transparent reporting form
- https://doi.org/10.7554/eLife.41676.017





