Chemical Informatics: Sense of achievement
The question of how we and other animals perceive the surrounding world was tackled by Aristotle more than 2300 years ago. Since then we have gained quite a good understanding of visual perception. Humans and most other animals employ a small number of different types of visual receptor, each being most sensitive to light of a specific wavelength and less sensitive to shorter or longer wavelengths (Schnapf et al., 1987). Using different receptor types, with overlapping sensitivity ranges, we can detect light with wavelengths between about 380 nm and 750 nm. Hearing is also well understood: sounds of different wavelengths activate different types of sensory neurons to provide coverage over a range of wavelengths (Masterto et al., 1969). However, the way that we respond to our chemical environment—that is, the way we respond to different smells and tastes—is much more complicated.
Contrary to vision and audition, olfaction has to deal with cues that are not arranged along a linear scale. The nose is exposed to several hundred thousand odorants that differ in chemical structure and in ecological relevance. One might imagine that the nose would need numerous different receptor types—each type sensitive to just a single compound—to detect and discriminate all the relevant odorants. However, as always, evolution found a smarter way. As first discovered by Richard Axel and Linda Buck in 1991—and rewarded with a Nobel Prize in 2004—animals are equipped with a relatively small, species specific, number of olfactory receptors (Buck and Axel, 1991): mice have more than 900 types, humans about 400, and the vinegar fly D. melanogaster has around 60.
Only a few, very important odorants—such as pheromones (Nakagawa et al., 2005) or the odorants given off by rotten food (Stensmyr et al., 2012)—have a one-to-one relationship with specific olfactory receptors. In general, a single receptor can detect a range of different odorants, and a single odorant can target a range of receptors, with a given odorant being identified through the pattern of receptors that it activates (Hallem and Carlson, 2006). It is thought that this so-called combinatorial olfactory code is employed by insects and also by vertebrates (Vosshall, 2000; Kauer and White, 2001). However, many of the details of the interactions between the odorant molecules and the receptors remain mysterious. Why, for example, do odorants with similar structures sometimes target different receptors, whereas other odorants with clearly different structures often target the same receptor.
Now, in eLife, Sean Boyle, Shane McInally and Anandasankar Ray of the University of California at Riverside describe a new method that can predict which odorants interact with which receptors much more accurately than previous methods (Boyle et al., 2013). During the last decade many groups have screened the sensory range of the odorant receptors of the vinegar fly, and a total of 251 different odorants are known to be able to activate at least one receptor. Although this is a tiny number compared with the number of odorants that flies are usually exposed to, Boyle, McInally and Ray were able to gain fresh insights into the receptor-odorant interactions by performing a highly detailed meta-analysis on these 251 odorants to identify the properties that cause an odorant to target a particular receptor (Figure 1). In addition to the ‘usual suspects’ of molecular properties (e.g., whether the odorant is an alcohol, an ester or an aldehyde), they took into account some 3,224 physical and/or chemical properties of the odorants, including obvious properties like molecular weight and three-dimensional structure, and less obvious properties like the ‘eigenvalue sum from electronegativity weighted distance matrix’.
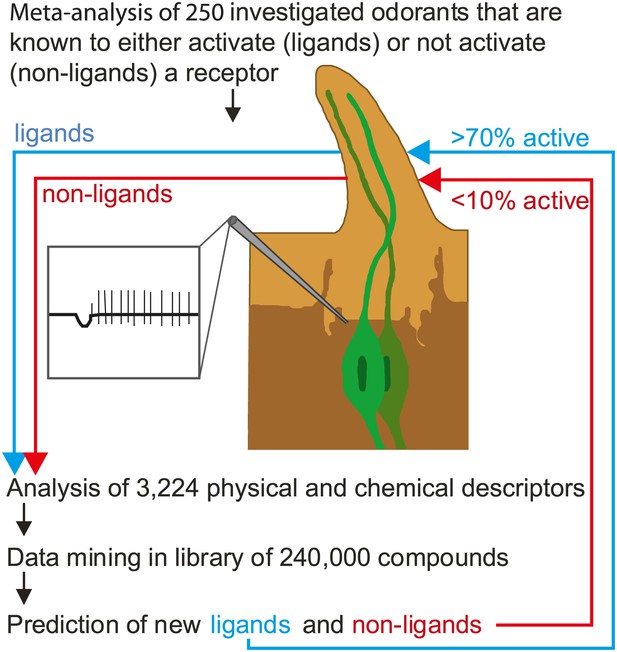
Predicting odorant-receptor interactions.
Boyle et al. performed a meta-analysis of 250 odorants and 51 receptors and developed an algorithm (based on some 3,224 physical and chemical properties of the odorants) to predict whether a given odorant will interact with a given receptor. This algorithm was then used to ‘mine’ a library of 240,000 compounds and identify ligands (blue line) and non-ligands (red line) for nine receptors. Experiments were performed with 141 compounds (11–23 per receptor): 71% of the compounds that were predicted to be ligands were found to interact with the relevant receptor, and less than 10% of the compounds that were predicted to be non-ligands were found to interact. The illustration shows an insect sensillum housing two olfactory receptor neurons (one pale green, the other dark green), each with a cell body and a nucleus, and a dendrite that extends into the tip of the sensillum. The tip is filled with a fluid called the sensillum lymph (pale brown) that is excreted by trichogen cells (dark brown). The expanded detail shows the neuronal response to a ligand as measured in the single sensillum recordings performed by Boyle et al.
This approach was pioneered by groups at Goethe University in Frankfurt (Schmuker et al., 2007) and the Weizmann Institute (Haddad et al., 2008). However, instead of analysing all the receptors and all the physical and chemical properties, the Riverside researchers used an algorithm that allowed the most critical properties for each receptor to be identified. Next they screened a list of more of 240,000 odorants to find those that they expected to interact with nine different receptors. Finally, they tested these predictions in experiments: Their predictions were correct more than 70% of the time, compared with a success rate of just 10% for odorants chosen at random. Hence, although odorants do not follow any linear rules like light and sound, we can still use their physical and chemical properties to predict whether an odorant interacts with a specific receptor and later, we hope, be able to understand why it interacts.
These results will be of interest beyond a narrow group of specialists. According to the United Nations Food and Agriculture Organization, insects and insect-spread diseases are responsible for an estimated 20–40% of world-wide crop production being lost every year. Furthermore, malaria and dengue fever, which are both spread by mosquitoes, kill more than 1 million people every year (and infect another 250 million). As insects typically use olfactory cues to find new hosts, a better understanding of odorant-receptor interactions promises substantial improvements for human food supply and health.
References
-
Imaging and coding in the olfactory systemAnnu Rev Neurosci 24:963–979.https://doi.org/10.1146/annurev.neuro.24.1.963
-
Olfaction in DrosophilaCurr Opin Neurobiol 10:498–503.https://doi.org/10.1016/S0959-4388(00)00111-2
Article and author information
Author details
Publication history
- Version of Record published: October 15, 2013 (version 1)
Copyright
© 2013, Knaden and Hansson
This article is distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use and redistribution provided that the original author and source are credited.
Metrics
-
- 605
- views
-
- 49
- downloads
-
- 1
- citations
Views, downloads and citations are aggregated across all versions of this paper published by eLife.
Download links
Downloads (link to download the article as PDF)
Open citations (links to open the citations from this article in various online reference manager services)
Cite this article (links to download the citations from this article in formats compatible with various reference manager tools)
Further reading
-
- Neuroscience
Combining information from multiple senses is essential to object recognition, core to the ability to learn concepts, make new inferences, and generalize across distinct entities. Yet how the mind combines sensory input into coherent crossmodal representations - the crossmodal binding problem - remains poorly understood. Here, we applied multi-echo fMRI across a four-day paradigm, in which participants learned 3-dimensional crossmodal representations created from well-characterized unimodal visual shape and sound features. Our novel paradigm decoupled the learned crossmodal object representations from their baseline unimodal shapes and sounds, thus allowing us to track the emergence of crossmodal object representations as they were learned by healthy adults. Critically, we found that two anterior temporal lobe structures - temporal pole and perirhinal cortex - differentiated learned from non-learned crossmodal objects, even when controlling for the unimodal features that composed those objects. These results provide evidence for integrated crossmodal object representations in the anterior temporal lobes that were different from the representations for the unimodal features. Furthermore, we found that perirhinal cortex representations were by default biased towards visual shape, but this initial visual bias was attenuated by crossmodal learning. Thus, crossmodal learning transformed perirhinal representations such that they were no longer predominantly grounded in the visual modality, which may be a mechanism by which object concepts gain their abstraction.
-
- Genetics and Genomics
- Neuroscience
Genome-wide association studies have revealed >270 loci associated with schizophrenia risk, yet these genetic factors do not seem to be sufficient to fully explain the molecular determinants behind this psychiatric condition. Epigenetic marks such as post-translational histone modifications remain largely plastic during development and adulthood, allowing a dynamic impact of environmental factors, including antipsychotic medications, on access to genes and regulatory elements. However, few studies so far have profiled cell-specific genome-wide histone modifications in postmortem brain samples from schizophrenia subjects, or the effect of antipsychotic treatment on such epigenetic marks. Here, we conducted ChIP-seq analyses focusing on histone marks indicative of active enhancers (H3K27ac) and active promoters (H3K4me3), alongside RNA-seq, using frontal cortex samples from antipsychotic-free (AF) and antipsychotic-treated (AT) individuals with schizophrenia, as well as individually matched controls (n=58). Schizophrenia subjects exhibited thousands of neuronal and non-neuronal epigenetic differences at regions that included several susceptibility genetic loci, such as NRG1, DISC1, and DRD3. By analyzing the AF and AT cohorts separately, we identified schizophrenia-associated alterations in specific transcription factors, their regulatees, and epigenomic and transcriptomic features that were reversed by antipsychotic treatment; as well as those that represented a consequence of antipsychotic medication rather than a hallmark of schizophrenia in postmortem human brain samples. Notably, we also found that the effect of age on epigenomic landscapes was more pronounced in frontal cortex of AT-schizophrenics, as compared to AF-schizophrenics and controls. Together, these data provide important evidence of epigenetic alterations in the frontal cortex of individuals with schizophrenia, and remark for the first time on the impact of age and antipsychotic treatment on chromatin organization.





