Cell Signaling: Extraordinary effects of unnatural pairings
As with John Lennon and Paul McCartney, pairings can sometimes exceed the sum of their parts. Many biological processes also rely on proteins forming pairs: for example, a wide variety of cells (particularly those involved in the immune system) release small proteins called cytokines to communicate with other cells, and typically it takes two cytokine receptors to detect one of these proteins. Now, in eLife, Christopher Garcia and colleagues at Stanford University School of Medicine – including Ignacio Moraga as first author – report that they have developed a new class of molecule called synthekines that bind to different pairs of cytokine receptors than the cytokines they are based on (Moraga et al., 2017).
Each cytokine interacts with a unique cytokine receptor pair and activates signaling pathways that lead to changes in gene expression (Figure 1). In mammals, there are ∼40 cytokine receptors that employ the Jak-STAT signaling pathway: this is an evolutionarily conserved pathway composed of four Jak proteins and seven STAT proteins that cooperate to sense when specific cytokine receptors form pairs (Villarino et al., 2017). Simple math tells us that, if all such receptors could freely associate with each other, there would be 40x40=1,600 combinations of pairs. However, this is not the case; the cytokines present in an organism determine which receptor pairings occur, limiting the actual number of pairings to less than 50.
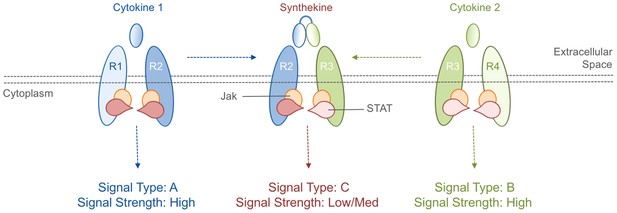
Cytokines and synthekines.
The cytokines present in an organism dictate which cytokine receptors can form pairs: in this schematic example, cytokine 1 causes receptors R1 and R2 to form a pair (left), while cytokine 2 causes receptors R3 and R4 to form a pair (right). Moraga et al. made new molecules called synthekines, with each synthekine being a composite of two mutant cytokines that each bind only one of their related receptors. These synthekines trigger the formation of ‘unnatural’ pairs of receptors (R2 and R3 in this case). The Jak-STAT signaling cascade downstream of the synthekine is weaker than the cascades triggered by the parent cytokines: there are also other differences in the signaling cascades (see main text).
Cytokines bind to a region of cytokine receptors known as the ligand-binding domain, which sticks out from the cell surface. This causes the signaling domains of two receptors, which are inside the cell, to interact with one another and activate Jak-STAT signaling. Moraga et al. first tested whether ‘unnatural’ cytokine receptor pairings can trigger Jak-STAT signaling. To that end, they devised an experimental system in which the signaling domains of 10 different cytokine receptors were each fused to one of two ligand-binding domains. These ligand-binding domains were compatible with a single cytokine that, in turn, could induce any one of the 10x10=100 possible pairings of signaling domains, depending on which two were fused to the ligand-binding domains.
Moraga et al. discovered that most combinations of ligand-binding and signaling domains were able to activate Jak-STAT signaling. This confirms that the pairing of cytokine receptors is sufficient to trigger signaling inside cells and that the specificity of the interaction between cytokines and ligand-binding domains is what determines which receptors can form pairs. The researchers also showed that a cytokine receptor can even trigger signaling when forced to form a pair with a completely different class of receptor molecule, such as a receptor tyrosine kinase.
Moving beyond proof-of-principle, Moraga et al. developed a way to produce ‘unnatural’ receptor pairings in native human cells. Based on the biophysics of the interactions between cytokines and their receptors, they designed mutant versions of three cytokines that were capable of binding to only one component of their ‘natural’ receptor pairs. Two of these mutant cytokines were then linked together to create synthetic cytokines (named synthekines) that can cause one receptor for each mutant cytokine to form a pair that would not naturally occur (Figure 1).
The experiments show that Jak-STAT signaling activity downstream of two synthekines called SY1 and SY2 were distinct from those of their parent cytokines. The responses driven by synthekines generally had lower overall levels of Jak-STAT activity compared to the responses triggered by their parent cytokines. Furthermore, the patterns of STAT protein activity differed. For example, STAT1 is the most active STAT protein in the response to SY2, but least active in the response to its parent cytokines. Furthermore, a technique called high-dimensional mass cytometry revealed that other signaling pathways, particularly downstream of a protein called phosphoinositide 3-kinase, were altered. Thus, synthekines are not just a watered down combination of their parent cytokines but, instead, are new biological entities with unique signaling properties. This makes them useful research tools and promising drug candidates.
There are clear precedents for the current work (see, for example, Cunningham and Wells, 1991; Shanafelt et al., 2000; Levin et al., 2012; Junttila et al., 2012; Levin et al., 2014; Moraga et al., 2015; Ng and Galipeau, 2015). However, by presenting an expansive, multi-pronged methodology for devising and testing chimeric cytokines, Moraga et al. move the field on significantly. Furthermore, they introduce new design principles to circumvent some of the problems encountered by previous efforts; for instance, they report how to prevent the individual components of the synthekines from interacting with their ‘natural’ receptor pairs.
Of course, like all pioneering endeavors, the studies of Moraga et al. raise questions and compel further investigations. The cellular and molecular consequences of synthekines (versus their parent cytokines) have only begun to be explained, and their bioactivity in animals, particularly regarding therapeutic uses, has yet to be addressed. The latter will require the generation of synthekines that are compatible with rodents and other animal models because the current versions are made from human constituents. However, there appear to be no major obstacles to doing this.
All told, this work stands as a triumph of scientific imagination, a lucid illustration that we need not be constrained by the rules of engagement prescribed within the genome. After all, given the ever-growing practicality of synthetic biology, we are living in a time where, in the words of Lennon and McCartney, there's “nothing you can make that can't be made” (Lennon and McCartney, 1967).
References
-
Redirecting cell-type specific cytokine responses with engineered interleukin-4 superkinesNature Chemical Biology 8:990–998.https://doi.org/10.1038/nchembio.1096
-
Magical Mystery TourAll You Need Is Love, Magical Mystery Tour, London, EMI Records.
-
Mechanisms and consequences of Jak-STAT signaling in the immune systemNature Immunology 18:374–384.https://doi.org/10.1038/ni.3691
Article and author information
Author details
Publication history
- Version of Record published: May 12, 2017 (version 1)
- Version of Record updated: May 19, 2017 (version 2)
Copyright
This is an open-access article, free of all copyright, and may be freely reproduced, distributed, transmitted, modified, built upon, or otherwise used by anyone for any lawful purpose. The work is made available under the Creative Commons CC0 public domain dedication.
Metrics
-
- 1,204
- views
-
- 188
- downloads
-
- 1
- citations
Views, downloads and citations are aggregated across all versions of this paper published by eLife.
Download links
Downloads (link to download the article as PDF)
Open citations (links to open the citations from this article in various online reference manager services)
Cite this article (links to download the citations from this article in formats compatible with various reference manager tools)
Further reading
-
- Cell Biology
- Structural Biology and Molecular Biophysics
Mutations in the human PURA gene cause the neurodevelopmental PURA syndrome. In contrast to several other monogenetic disorders, almost all reported mutations in this nucleic acid-binding protein result in the full disease penetrance. In this study, we observed that patient mutations across PURA impair its previously reported co-localization with processing bodies. These mutations either destroyed the folding integrity, RNA binding, or dimerization of PURA. We also solved the crystal structures of the N- and C-terminal PUR domains of human PURA and combined them with molecular dynamics simulations and nuclear magnetic resonance measurements. The observed unusually high dynamics and structural promiscuity of PURA indicated that this protein is particularly susceptible to mutations impairing its structural integrity. It offers an explanation why even conservative mutations across PURA result in the full penetrance of symptoms in patients with PURA syndrome.
-
- Microbiology and Infectious Disease
- Structural Biology and Molecular Biophysics
African trypanosomes replicate within infected mammals where they are exposed to the complement system. This system centres around complement C3, which is present in a soluble form in serum but becomes covalently deposited onto the surfaces of pathogens after proteolytic cleavage to C3b. Membrane-associated C3b triggers different complement-mediated effectors which promote pathogen clearance. To counter complement-mediated clearance, African trypanosomes have a cell surface receptor, ISG65, which binds to C3b and which decreases the rate of trypanosome clearance in an infection model. However, the mechanism by which ISG65 reduces C3b function has not been determined. We reveal through cryogenic electron microscopy that ISG65 has two distinct binding sites for C3b, only one of which is available in C3 and C3d. We show that ISG65 does not block the formation of C3b or the function of the C3 convertase which catalyses the surface deposition of C3b. However, we show that ISG65 forms a specific conjugate with C3b, perhaps acting as a decoy. ISG65 also occludes the binding sites for complement receptors 2 and 3, which may disrupt recruitment of immune cells, including B cells, phagocytes, and granulocytes. This suggests that ISG65 protects trypanosomes by combining multiple approaches to dampen the complement cascade.






