An essential role for maternal control of Nodal signaling
Figures
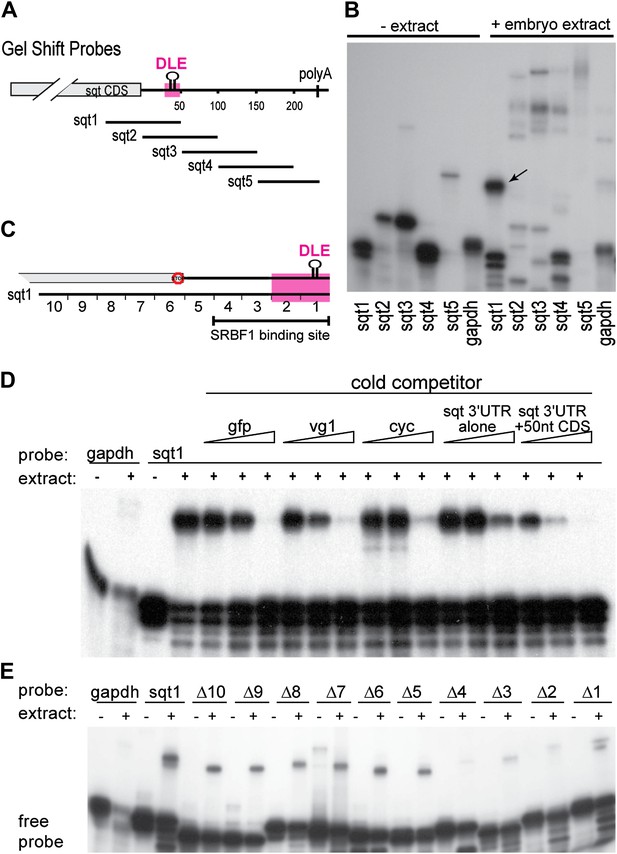
SRBF1 binds the sqt Dorsal Localization Element (DLE).
(A) Schematic of overlapping 100 nucleotide radioactive RNA gel-shift probes spanning the sqt 3′UTR. Position of DLE is highlighted in magenta. (B) Autoradiogram showing sqt 3′UTR probes incubated with embryo extract. Several binding activities were observed on the various probes. The ‘sqt RNA Binding Factor 1’ (SRBF1; black arrow) shift, is detected on the DLE-containing sqt1 probe, and not on other probes. (C) Schematic showing the SRBF1 binding site. sqt DLE is highlighted in magenta and the red octagon indicates the stop codon. (D) Competition gel-shift assay shows that SRBF1 binds specifically to sqt RNA. The sqt 3′UTR with 50 nucleotides of coding sequence competes more strongly than control gfp, vg1 or cyclops (cyc) RNA for binding to sqt1 probe. Triangles represent 4-fold increases (from 5 ng to 80 ng) of cold competitor RNA. Thus, SRBF1 preferentially binds DLE-sequences. (E) The SRBF1 binding site overlaps the DLE. RNA gel-shifts were performed with the sqt1 10 nt deletion series.
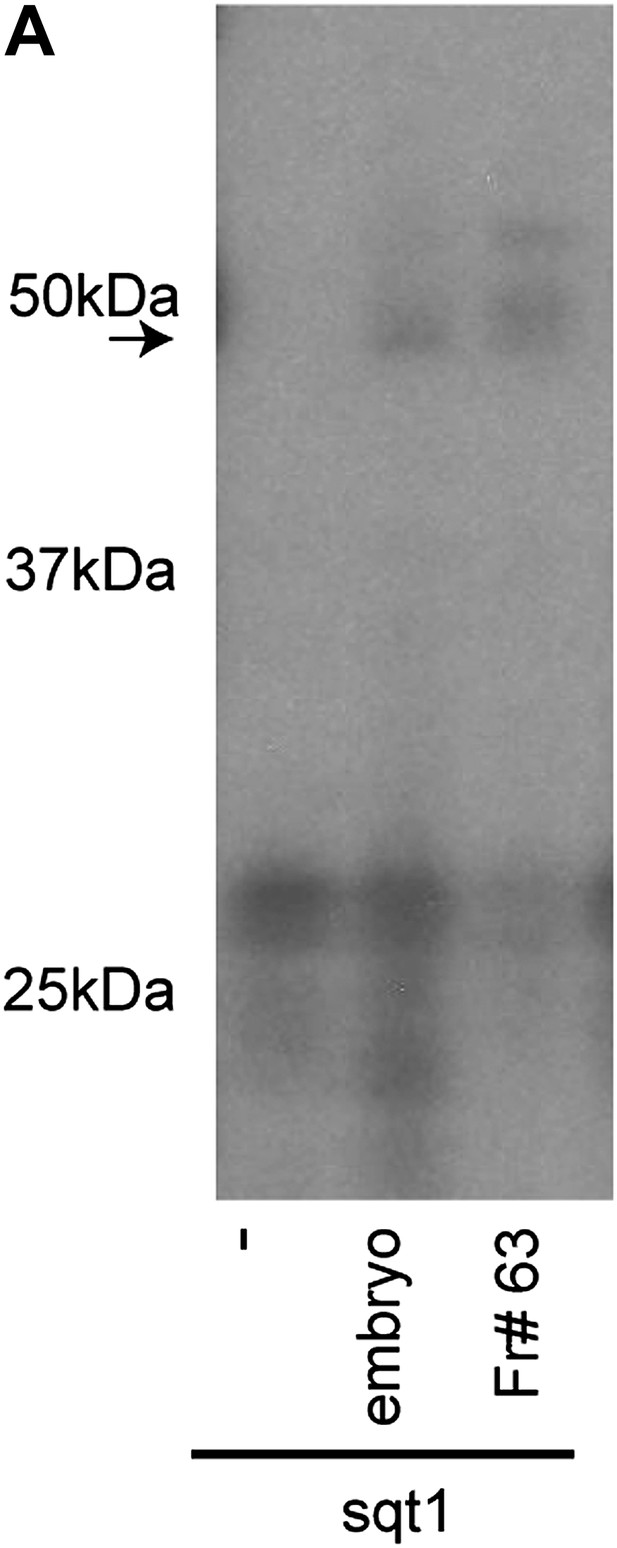
SRBF1 is an ∼50 kDa protein.
(A) RNA cross-linking autoradiogram showing that an ∼50 kDa UV cross-linking activity present in fraction#63 is similar to total embryo extracts. The ∼50 kDa peptide was excised from a native PAGE gel, and identified by mass spectrometry.
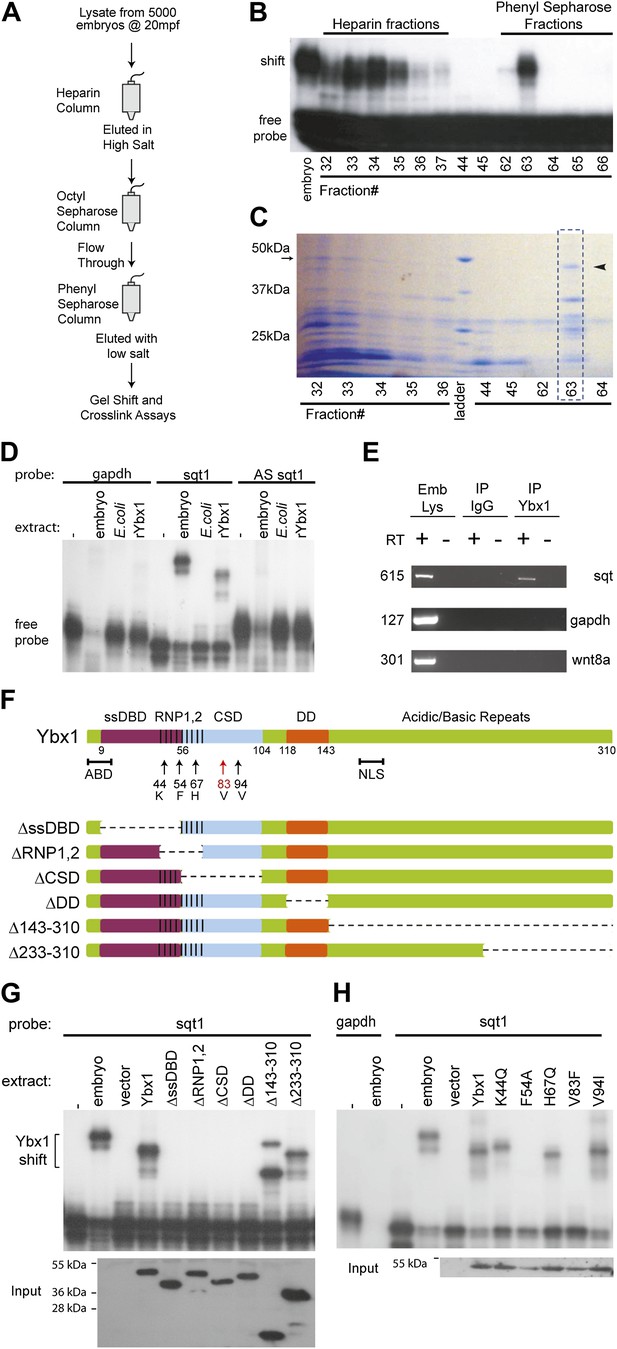
SRBF1 contains the nucleic acid binding protein Ybx1.
(A) Extracts from 5000 embryos collected at 20 mpf were sequentially fractionated on multiple chromatography columns, until SRBF1 was partially pure. At each stage, fractions containing SRBF1 activity were pooled, and loaded onto the next column for further purification. (B) A representative native PAGE gel showing SRBF1 purification. Gel-shift analysis of fractions from the heparin and phenyl sepharose columns show SRBF1 activity in fractions 32–37 from heparin and fractions 62–63 from the phenyl sepharose columns. Fractions 33–35 were pooled and added to the hydrophobic columns. Fraction 63 from the phenyl sepharose column contains partially purified SRBF1. (C) A Coomassie-blue stained SDS-PAGE gel of the fractions in B show a ∼48 kDa band that co-fractionates with SRBF1 (black arrowhead in fraction#63). The 48 kDa band from fraction#63 was excised, sequenced by mass spectrometry, and found to contain Ybx1 peptides. (D) Gel-shift analysis shows recombinant Ybx1 (rYbx1), similar to embryonic SRBF1, binds to sqt1, but not to control gapdh or antisense sqt1 probes. (E) Ybx1 binds sqt RNA in vivo. RT-PCR shows sqt RNA but not control gapdh or wnt8a RNA in RNA-IP with αYbx1 antibodies. Control IgG antibodies do not show any RT-PCR product. RT-PCR from whole embryo lysates is the positive control. PCR product sizes are indicated on the left. (F) Schematic diagram showing domain structure of wild-type and mutant Ybx1 proteins. The position of amino acid substitutions is indicated by arrows (V83 in red and all other residues in black). Deletions are indicated by dashed lines. The actin binding domain (ABD), single stranded DNA-binding domain (ssDBD; magenta), RNA-binding domains 1 and 2 (RNP1,2; hashed black lines), Cold shock domain (CSD, blue), dimerization domain (DD; orange), and nuclear localization sequence (NLS) are shown; numbers indicate the amino acid residue. (G) Domain analysis of Ybx1. The nucleic acid binding domain (ssDBD, magenta bar in F; CSD, blue bar in F; RNP1,2, hashed lines in F) is required for binding to sqt1, as is the dimerization domain (DD, orange bar in F). In contrast, the C-terminus of Ybx1 (Δ143–310 and Δ233–310) is dispensable for binding to sqt1. A western blot with α-His tag antibodies shows expression of the mutant Ybx1 proteins. (H) Point mutations in Ybx1 identify key amino acid residues that confer sqt RNA binding. K44, F54, and H67 are expected to contact RNA based upon NMR structure prediction. F54A abolishes binding, whereas H67Q does not affect binding at the protein concentrations shown. V83F abolishes sqt1 binding, whereas V94I binds sqt1. Western blot with α-His tag antibodies shows expression of mutant Ybx1 proteins.
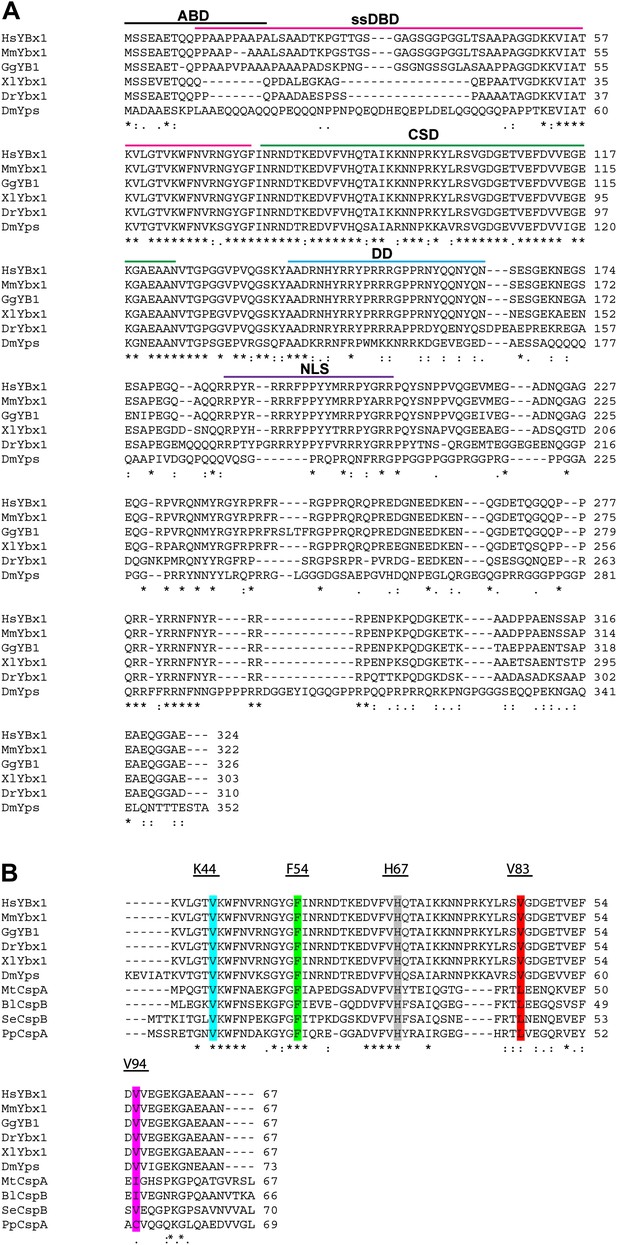
Alignment of Ybx1 sequences shows conservation across species.
(A) Alignment of Ybx1 sequences indicating the actin binding domain (ABD), single stranded DNA-binding domain (ssDBD), cold shock domain (CSD), dimerization domain (DD), and nuclear localization sequence (NLS). Species names and Genbank Accession numbers are as follows: Homo sapiens, AAI06046.1; Mus musculus, AAH61634.1; Gallus gallus, NM_204414.1; Danio rerio, AAI68507.1; Xenopus laevis, AAH41191.1; Drosophila melanogaster, NM_079309.3. (B) Alignment of cold shock proteins from bacterial species with eukaryotic CSD-containing proteins. The K44, F54, and H67 highlighted residues were identified by NMR to contact RNA. The V83 and V94 residues that were mutated by ENU (identified by TILLING) are also highlighted. Species name and GenBank Accession numbers for bacterial proteins are Pseudomonas putida, ADR61621.1; Mycobacterium tuberculosis, CCE39069.1; Salmonella enterica, CAA72682.1; Bacillus licheniformis, AAU39879.1
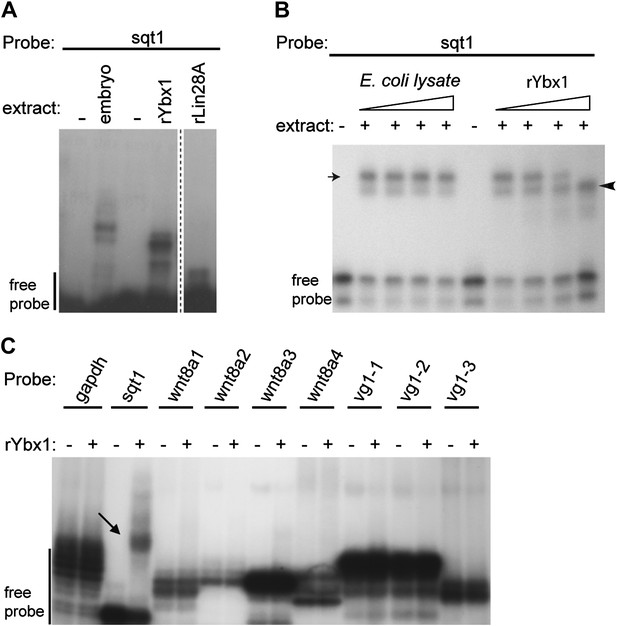
Ybx1 specifically binds to the sqt 3′UTR.
(A) Gel-shift assay showing that recombinant Ybx1 binds to sqt1 probe similar to embryo extracts, whereas recombinant Lin28A does not bind to sqt1. The rLin28A lane is from a different part of the same gel (demarcated by a dotted line). (B) Specificity of Ybx1 binding to sqt1. Gel-shift, showing that recombinant Ybx1 (black arrowhead) competes with native Ybx1 (arrow) for binding to the sqt1 probe. Triangles indicate fivefold increments of E. coli lysate or rYbx1. (C) Ybx1 binds sqt1 but not wnt8a, vg1 or gapdh probes. RNA gel-shift assay shows that only sqt1 probe shows a mobility shift with rYbx1 (black arrow) whereas probes spanning the wnt8 3′UTR (wnt8a 1-4) or vg1 3′UTR (vg1 1-3) do not show mobility shift with rYbx1.
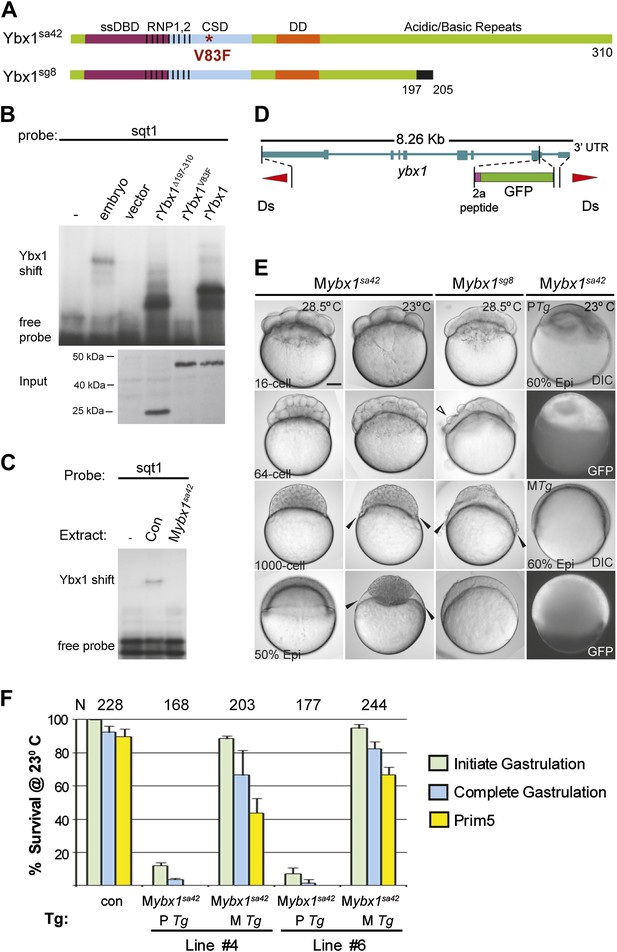
Maternal Ybx1 is essential for early development.
(A) Schematic of Ybx1 showing the various domains, the V83F mutation in the CSD in ybx1sa42, and deletion of residues 197–310 in ybx1sg8 mutants. Black box in Ybx1sg8 indicates frameshift after residue 197 and premature stop after residue 205. (B) rYbx1V83F lacks detectable DLE-binding activity similar to vector control, whereas rYbx1 and rYbx1Δ197–310 peptides, and embryo lysates show binding to sqt1 probes. Western blots to detect 6xHis epitope tags show expression of recombinant Ybx1 proteins. (C) Mybx1sa42 embryo extracts show no detectable binding to sqt1 probe compared to control extracts. (D) Schematic representation of the ybx1 genomic locus (blue) with positions of viral 2a peptide (magenta bar) and gfp (green box) indicated. Red triangles indicate Ds transposon terminal repeats. (E) DIC photomicrographs showing 16-cell, 64-cell, 1000-cell and 50% epiboly stage embryos. Mybx1sa42 embryos are viable at 28.5°C. Mybx1sg8 embryos cleave aberrantly after 16-cells (open arrowhead). Mybx1sa42 embryos at 23°C, and Mybx1sg8 embryos fail to initiate gastrulation, form syncytia (black arrowheads), and arrest. Zygotic Ybx1-GFP expression from PTg does not rescue gastrula arrest in Mybx1, whereas maternal Ybx1-GFP expression from MTg leads to normal gastrulation. Scale bar, 100 μm. (F) Histogram showing rescue of gastrulation and survival till prim5 stage of Mybx1 mutants at 23°C by two independent MTg lines (MTg #4 and MTg #6). Some embryos with zygotic expression of ybx1 (PTg) from both lines can initiate gastrulation, but none survive to prim5. Error bars show standard deviation from three experiments. Number of embryos is shown on top of the histogram.
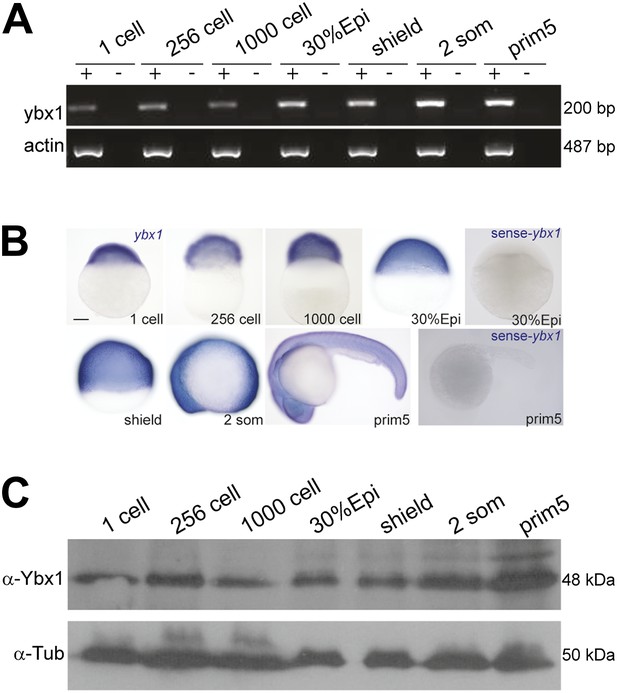
Expression of ybx1 RNA and Ybx1 protein in wild-type embryos.
(A) RT-PCR shows that ybx1 RNA is expressed at all stages examined. RT- (−) and actin PCRs were used as controls. (B) In situ hybridizations show that ybx1 RNA is not spatially restricted in embryos. Control sense probes show no expression. Scale bar, 100 μm. (C) Western blots to detect Ybx1 in embryo lysates show expression at all stages examined. Tubulin protein was detected as a control.
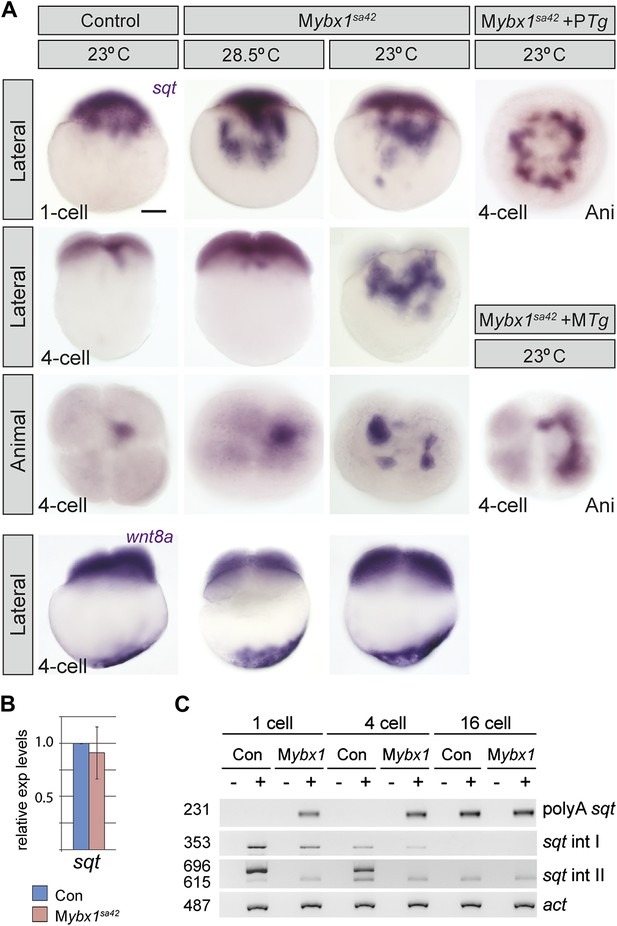
Ybx1 is required for localization and regulated processing of sqt RNA.
(A) Control embryos at 23°C show sqt RNA localization at the 1-cell and 4-cell stage. sqt RNA transport is delayed in 1-cell Mybx1sa42 embryos at 28.5°C, but localizes correctly by the 4-cell stage. At 23°C, sqt RNA largely remains in the yolk even at 4-cell stage and sqt RNA that reaches the blastoderm is mis-localized. Localization of sqt RNA is restored in Mybx1 by ybx1-2a-gfp MTg, but not by PTg. Localization of wnt8a RNA is normal in Mybx1sa42 mutants at 28.5°C and 23°C. Scale bar, 100 μm. (B) Q-RT-PCRs to detect total sqt RNA levels show a mild reduction in Mybx1 compared to controls. Error bars show standard deviation from three experiments. (C) RT-PCR to detect sqt in control and Mybx1 mutants at 1-cell, 4-cell and 16-cell stages. Products are indicated on the right, and sizes on the left. Polyadenylated sqt RNA is detected from 16-cells in controls, and at 1-cell in Mybx1 mutants. Splicing of sqt intron 1 and sqt intron 2 occurs earlier in Mybx1 embryos compared to controls. PCR to detect actin is shown as control.
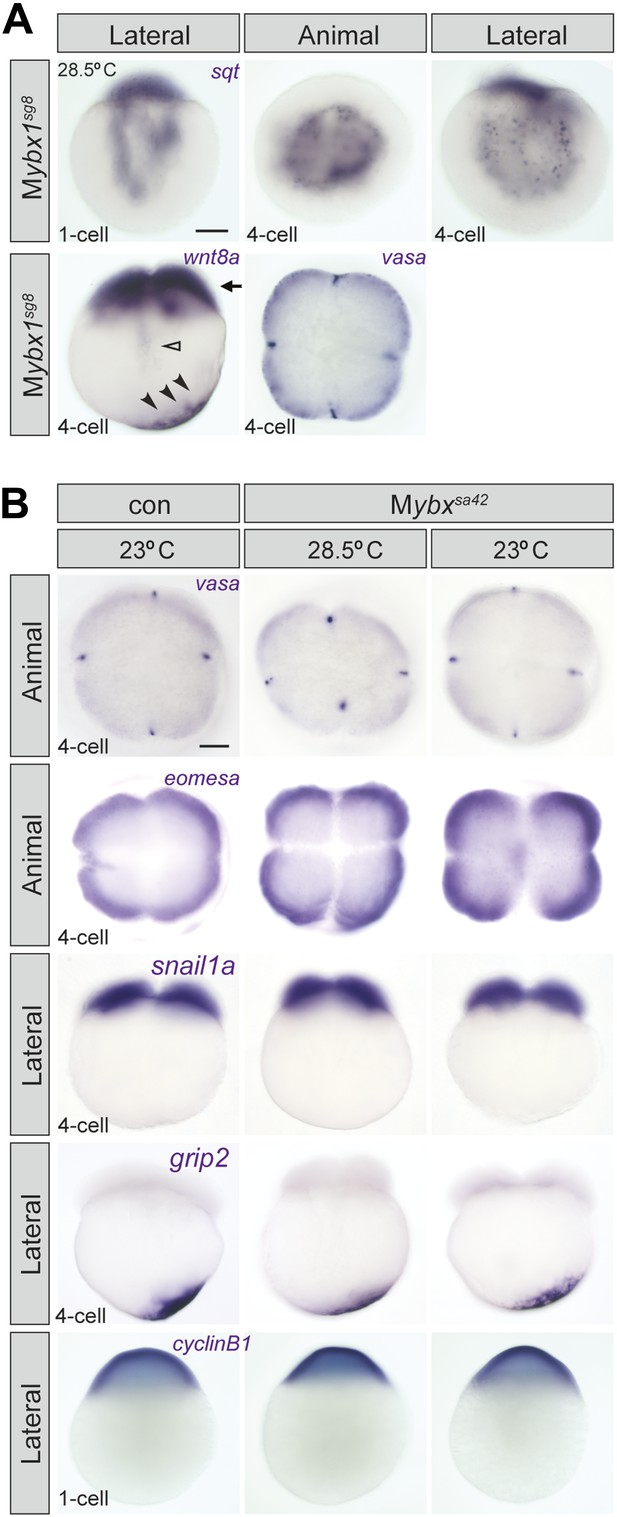
Localization of sqt RNA is affected in Mybx1 embryos, whereas vasa, eomesa, snail1a, grip2 and cyclin B1 are unaffected.
(A) Localization of sqt RNA is disrupted in Mybx1sg8 embryos at the 1-cell and 4-cell stage. Localization of wnt8a and vasa is generally unaffected in Mybx1sg8 (black arrowheads and black arrow). In a very small number of Mybx1sg8 embryos at the 4-cell stage, some wnt8a RNA is observed in the yolk (open arrowhead), reflecting a slight lag in transport. Scale bar, 100 μm. (B) Localization of maternal vasa, eomesa, snail1a, grip2 and cycin B1 transcripts is unaffected in Mybx1sa42 embryos at 28.5°C and 23°C.
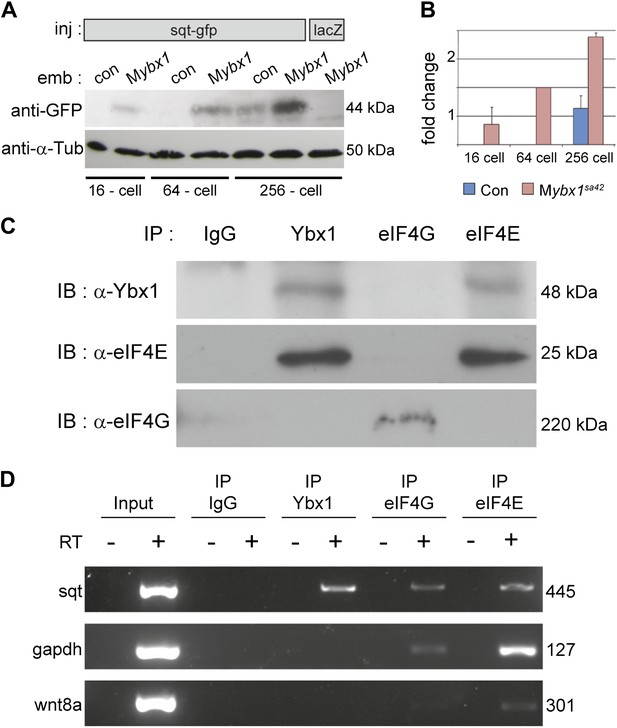
Ybx1 interacts with the translation initiation machinery and represses translation of sqt RNA.
(A) Western blot to detect GFP shows injected sqt-gfp is translated by 16-cells in Mybx1, whereas in controls, Sqt-GFP is detected at blastula stages, and lacZ control injection shows no Sqt-GFP. (B) Sqt-GFP protein expression is precocious and elevated in Mybx1 embryos. Error bars in B show standard deviation from three experiments. (C) Co-immunoprecipitation in embryo lysates followed by western blot analysis shows that Ybx1 interacts with eIF4E. eIF4G binds poorly with Ybx1. Faint smear in control IgG lane for eIF4G is spillover from input lane (see Figure 5—figure supplement 1C for complete blot for eIF4G). (D) Antibodies towards Ybx1, eIF4G and eIF4E pull down sqt RNA in embryos lysates. RT-PCR on the embryos lysates in panel C shows sqt RNA but not control gapdh or wnt8a RNA in RNA-IP with αYbx1 antibodies. Control IgG antibodies do not show any RT-PCR product, whereas antibodies to the translation initiation factor eIF4E can pull down sqt RNA, wnt8a and gapdh RNA, and antibodies to eIF4G detects weak bands for sqt and gapdh in the RNA-IPs. RT-PCR from whole embryo lysates is the positive control. PCR product sizes are indicated on the right.
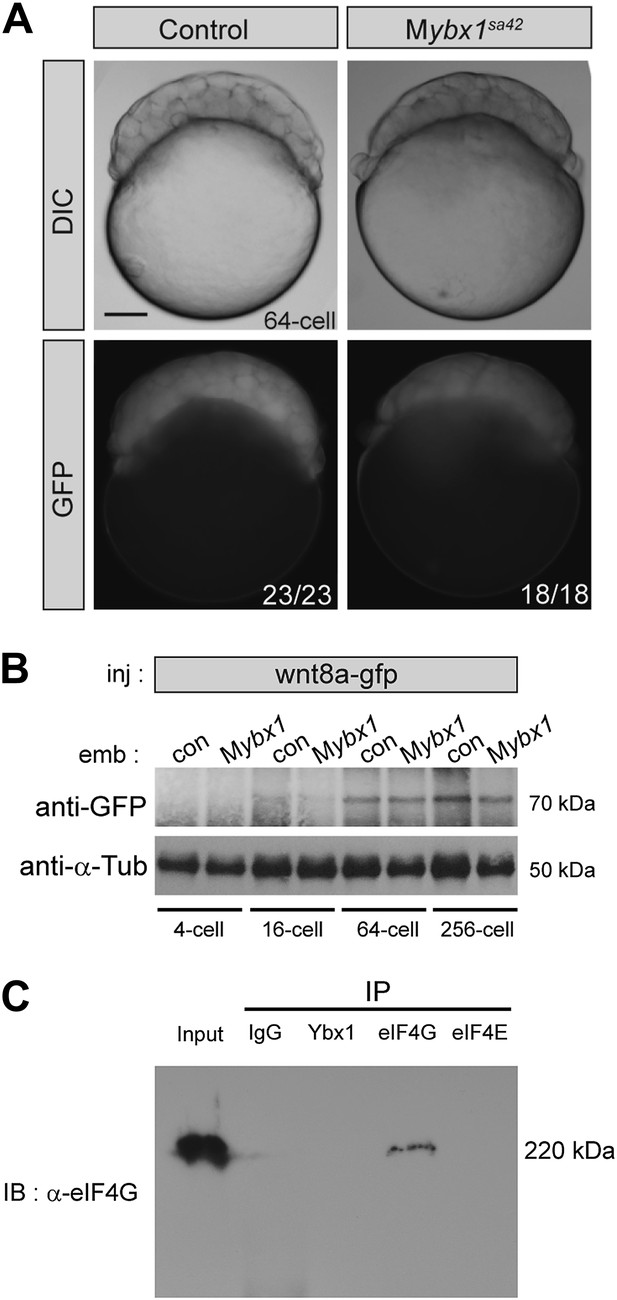
Translation of control RNAs is not affected in Mybx1 mutant embryos.
(A) Control GFP is not translated differentially in Mybx1 and control Pybx1 embryos. GFP RNA injected at the 1-cell stage is expressed similarly in 64-cell control and Mybx1 embryos. Scale bar, 100 μm. (B) Expression of Wnt8a-GFP protein (70 kDa) is similar in control and Mybx1 embryos at cleavage and early blastula stages. Tubulin expression was detected as control. (C) Western blot to detect eIF4G in co-immunoprecipitations from embryo lysates using antibodies to Ybx1, eIF4E and eIF4G shows a 220 kDa band only with eIF4G pull-down but not with Ybx1 or eIF4E. IgG antibodies were used as negative control and input embryo lysate is the positive control. A faint smear is observed in part of the IgG lane, due to spillover from the input.
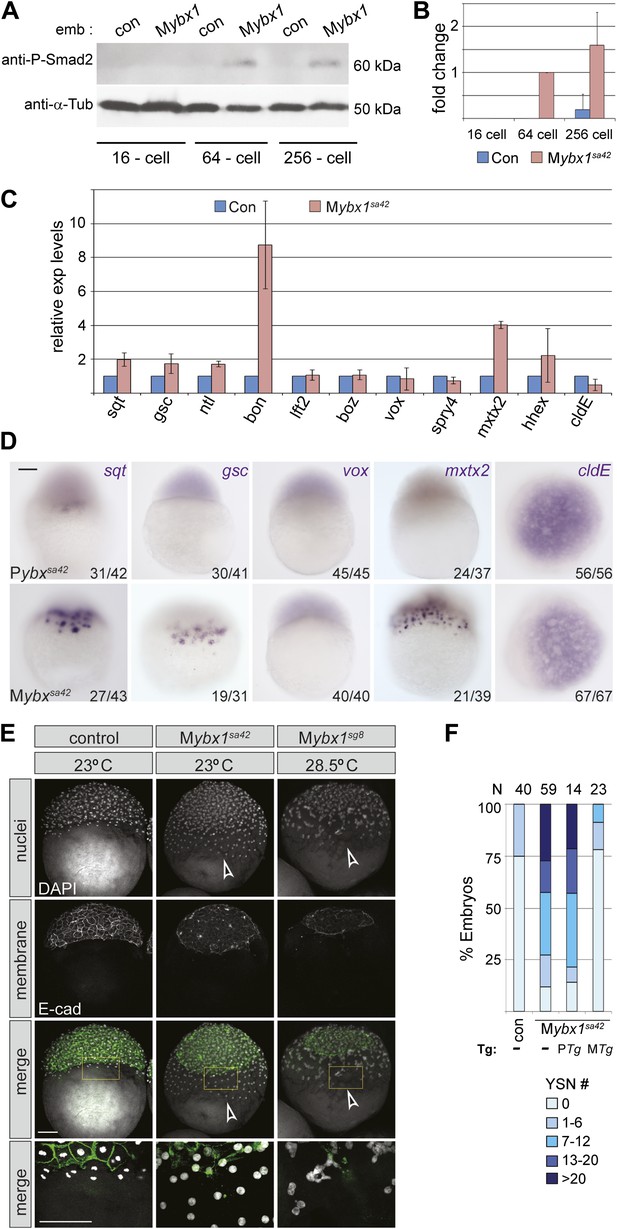
Nodal signaling is deregulated in Mybx1 embryos.
(A) Phosphorylated-Smad2 is detected at the 64-cell stage in Mybx1 embryos. (B) Phospho-Smad2 levels are elevated in Mybx1 embryos at cleavage stages, compared to controls. (C) Quantitative real-time RT-PCR shows that Nodal target (sqt, gsc, ntl, bon) and YSL gene expression (mxtx2) is elevated in Mybx1 compared to controls, whereas expression of lefty2, the Wnt target, boz, ventral mesoderm gene vox, FGF target spry4, and the enveloping layer (EVL) marker cldE, is either not significantly altered or marginally reduced. (D) Whole mount in situ hybridization shows expanded YSL domains of sqt, gsc, and mxtx2 in Mybx1 embryos; the cldE expression domain is not significantly different from controls, and vox is not detectable. Scale bar, 100 μm. (E) Mybx1 mutants have expanded YSL. DAPI staining to detect nuclei and E-cadherin immunostaining to detect membranes shows 1 tier of YSL nuclei (undergoing division) in control embryos. In Mybx1sa42 and Mybx1sg8 embryos, multiple tiers of YSL are observed (yellow boxed area). Bottom panels show higher magnification of yellow, boxed area. Cell membranes are clearly demarcated in control embryos, but appear fragmented in Mybx1 mutants. Scale bars, 100 μm. (F) Histogram showing YSL nuclei numbers in Mybx1 and control embryos, with or without ybx1 transgenes at 23°C. ∼75% of control embryos have no YSL nuclei and only 25% show 1–6 YSL nuclei, whereas ∼80% of Mybx1 embryos and Mybx1 embryos with PTg, show 7 or more YSL nuclei, and ∼25% show >20 YSL nuclei. Mybx1 embryos with ybx1 MTg show reduced numbers of YSL nuclei. Number of embryos scored is indicated above the histogram. Error bars in B and C show standard deviation from three experiments.
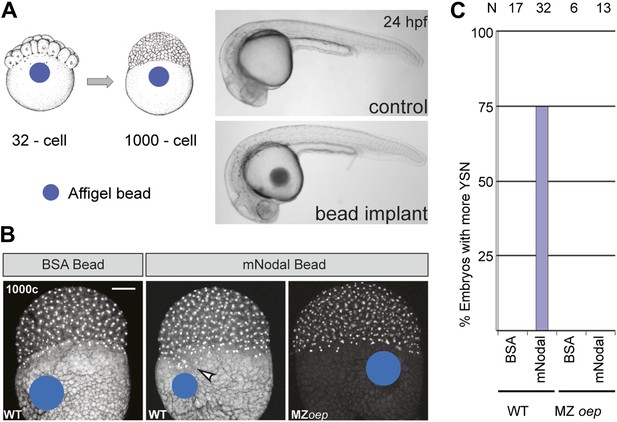
Excess Nodal protein from the yolk directly expands the extra-embryonic YSL.
(A) Affi-gel beads presoaked in either BSA or mNodal were implanted at the 32-cell stage and fixed at 1000-cell stage. BSA-bead implanted embryos develop normally and have no morphological defects at 24 hpf, similar to control embryos. (B) DAPI staining shows one tier of YSL nuclei in BSA bead-implanted wild-type embryos whereas mNodal bead-implanted embryos show many nuclei (arrowhead). MZoep embryos with Nodal beads are similar to control BSA bead-implanted embryos. Bead position indicated by blue dot. Scale bar, 100 μm. (C) Histogram showing percent wild-type or MZoep embryos with more YSN after bead implants.
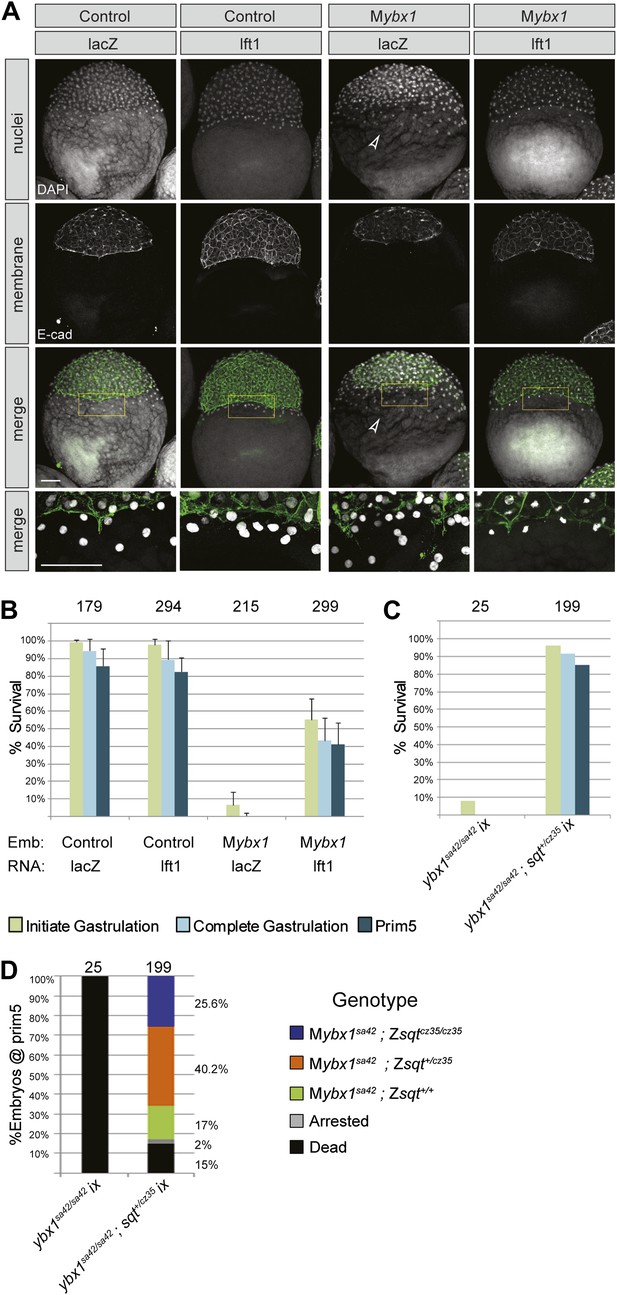
YSL and gastrulation defects in Mybx1 mutants can be rescued by blocking Nodal signaling.
(A) DAPI-stained nuclei and E-cadherin immunostained membranes are clearly demarcated in lacZ and lefty1 RNA-injected control embryos. E-cadherin staining appears fragmented and number of YSL nuclei is increased in Mybx1 mutant embryos injected with control lacZ RNA (white arrowhead). In Mybx1 embryos injected with lefty1 RNA, the number of YSL nuclei is restored to normal levels and membrane staining does not appear fragmented. Bottom panels show higher magnification of area boxed in yellow; scale bar, 100 μm. (B) Histogram showing rescue of gastrulation and survival till prim5 stage by injection of lefty1 RNA but not lacZ RNA in Mybx1 mutants that were subjected to temperature shift at 23°C. Error bars show standard deviation from three experiments. Number of embryos is shown on top of the histogram. (C) Histogram showing gastrulation and % survival in embryos from ybx1sa42/sa42;sqtcz35/+ crosses and Mybx1 mutants subjected to temperature shift at 23°C. Most embryos from ybx1sa42/sa42;sqtcz35/+ crosses initiate and complete gastrulation in comparison to Mybx1 sa42 mutants. (D) Histogram showing survival at 23°C and genotypes of embryos from matings of ybx1sa42/sa42;sqtcz35/+, in comparison to Mybx1sa42 embryos which do not survive at 23°C. The expected % for each genotype is 25% for Mybx1sa42;sqtcz35/cz35 and Mybx1sa42;sqt+/+, and 50% for Mybx1sa42;sqtcz35/+. All Mybx1sa42;sqtcz35/cz35 embryos (which have no Sqt signaling) survive, whereas many Mybx1sa42;sqtcz35/+ and Mybx1sa42;sqt+/+ do not survive at 23°C. Number of embryos scored is indicated above the histogram, and % observed for each genotype is indicated to the right of the colored bars.
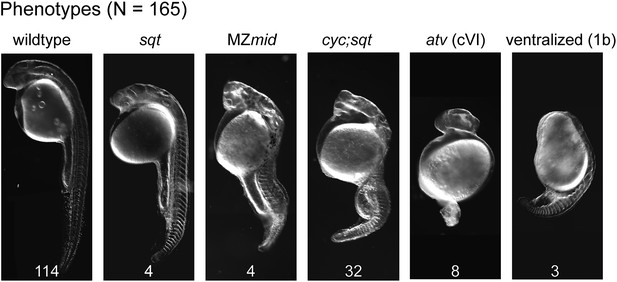
Mybx1;Zsqtcz35/cz35 mutant embryos show phenotypes typical of reduced Nodal activity.
DIC images of prim5 stage embryos from ybx1sa42/sa42;sqtcz35/+ intercrosses show a range of classic nodal phenotypes. Phenotypes scored (wild type-, squint-, MZmidway-, cyc;sqt-, antivin/lefty overexpression class VI-, and ventralized ichabod 1b-like) are indicated above the images, and numbers are given at the bottom of each image. Data compiled from two independent crosses.
Additional files
-
Supplementary file 1
Primer sequences.
- https://doi.org/10.7554/eLife.00683.018





