Brahma is essential for Drosophila intestinal stem cell proliferation and regulated by Hippo signaling
Figures
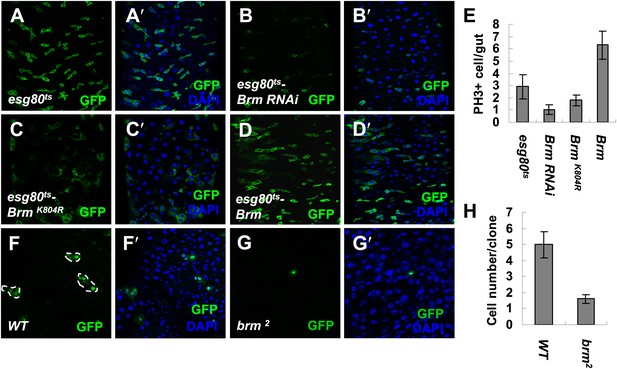
Brm is required for ISC proliferation in midguts.
(A–D′) Adult fly midguts expressing esg80ts-Gal4/UAS-GFP (esg80ts) (A and A′), Brm RNAi (esg80ts-Brm RNAi) (B and B′), esg80ts-Gal4/UAS-GFP-BrmK804R (esg80ts-BrmK804R) (C and C′) or esg80ts-Gal4/UAS-GFP-Brm (esg80ts-Brm) (D and D′) were immunostained with DAPI (blue). ISCs and EBs were marked by esgGal4-driven GFP expression. (E) Quantification of PH3+ cells of adult midguts of the indicated genotypes. The results represent the mean ± SEM, n = 10 for each genotype. (F–G′) Adult midguts containing nuclear localized GFP-labeled control MARCM clones (F and F′) or brm null allele brm2 clones (G and G′) were immunostained for DAPI (blue). Guts were dissected from the adult flies 72 hr after clone induction. (H) Quantification of the cell numbers of the control or mutant clones of the indicated genotypes. The results represent the mean ± SEM, n = 10 for each genotype. See also Figure 1—figure supplements 1 and 2.
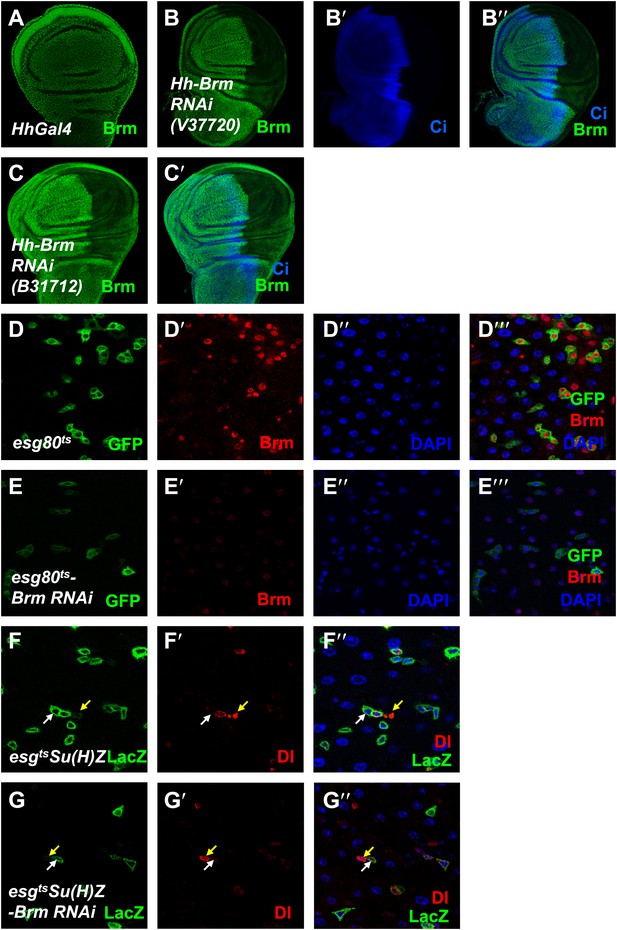
Brm is required for ISC proliferation.
(A–C′) The efficiency of Brm antibody was tested by immunostaining of the endogenous Brm in wild-type wing discs (A) or in the discs expressing Brm RNAi V37720 (B–B′′) and B31712 (C and C′) in the posterior compartment using hhGal4 driver. Discs were immunostained for Brm (green) and Ci (blue). (D–E′′′) Adult flies expressing esg80ts (D–D′′′), esg80ts-Brm RNAi (E–E′′′) were cultured at 29°C for 7 days. Midguts were dissected and immunostained for Brm (red) and DAPI (blue). (F–G′′) Flies of Su(H)Z controls (F–F′′) or flies expressing Brm RNAi in the ISCs/EBs (G–G′′) were cultured at 29°C for 7 days. Dl is detected by immunostaining (red). Su(H)-lacZ staining identifies the EBs with elevated Notch signaling (green). Cells that retain ISC identity (small nuclei, Dl positive and lacZ-negative) are indicated by yellow arrows, and EBs are indicated by white arrows.
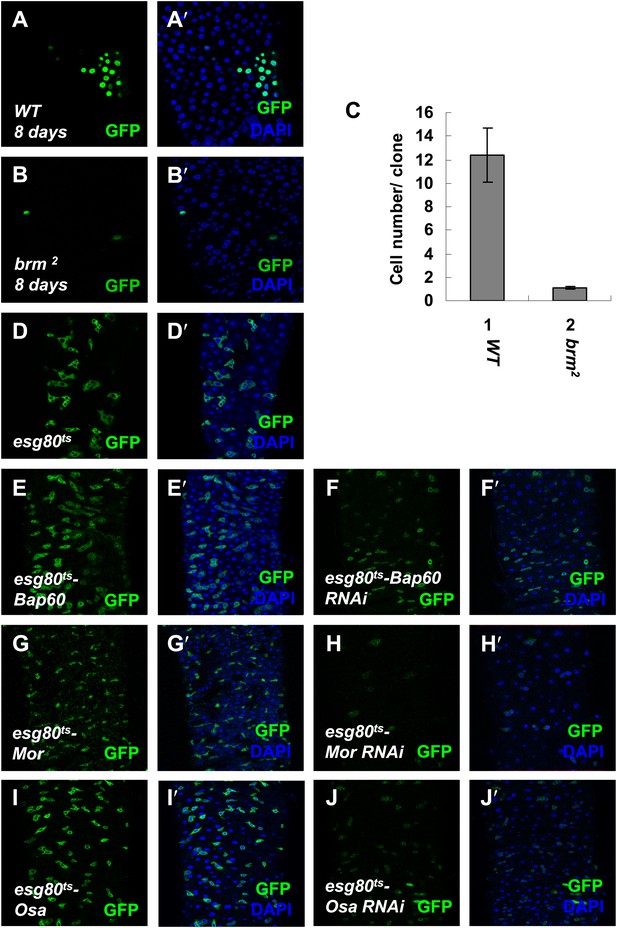
Brm complex is required for ISC proliferation.
(A–B′) Adult midguts containing nuclear localized GFP-labeled control MARCM clones (A and A′) or brm null allele brm2 clones (B and B′) were immunostained with DAPI (blue). Guts were dissected from the adult flies 8 days after clone induction. (C) Quantification of the cell numbers of the indicated control clones or mutant clones. The results represent the mean ± SEM, n > 10 for each genotype. (D–J′) Subunits of Brm complex function in ISC proliferation. Adult midguts expressing Bap60 (E and E′), Bap60 RNAi (NIG 4303R-1, F and F′), Mor (G and G′), Mor RNAi (VDRC 6969, H and H′), Osa (I and I′) and Osa RNAi (VDRC 7810, J and J′) with esg80ts driver were immunostained with DAPI (blue).
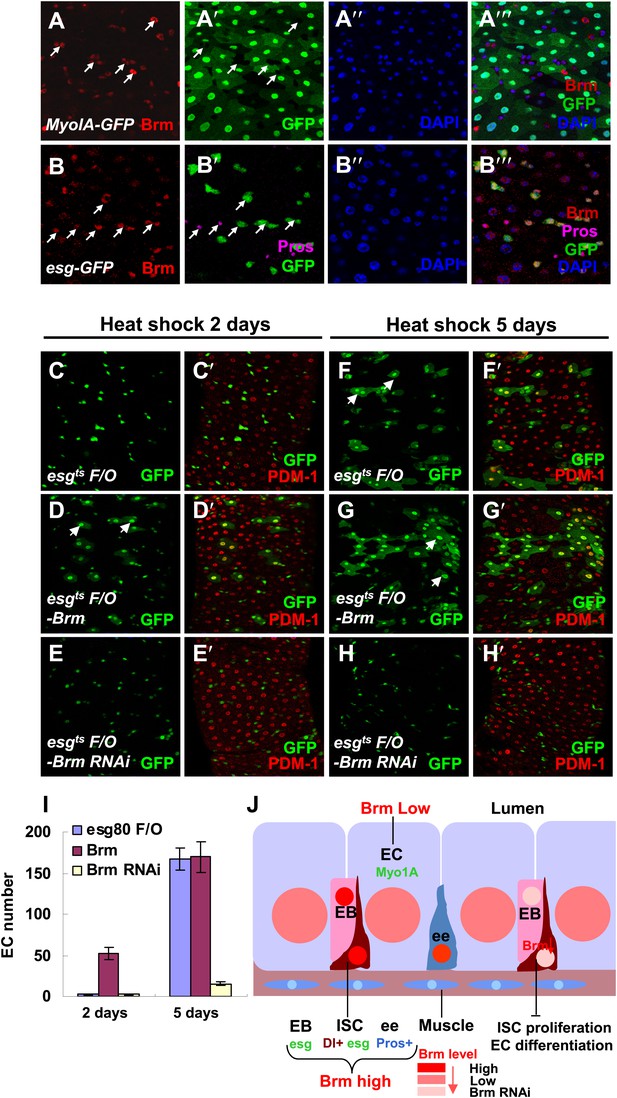
Brm is required for EC differentiation.
(A–B′′′) Adult guts of wild-type Myo1A-Gal4/UAS-GFP;tubGal80ts (A–A′′′) and esgGal4/UAS-GFP (B–B′′′) were immunostained with Brm antibody (indicated with arrows) to show the endogenous Brm protein level in the different cell types. (C–H′) Adult female midguts differentiation measured via the esgts F/O system. Transgenes were induced for 2 days (C–E′) or 5 days (F–H′). esgts F/O-Brm (D, D′ and G, G′) promoted the formation of ECs, while esgts F/O-Brm RNAi (E, E′ and H, H′) blocked the EC differentiation. ECs are marked by PDM-1 (red) and arrows. (I) Female posterior midguts were scored for GFP+ and PDM-1+ EC cells in the same region near the Malpighian tubules. The results represent the mean ± SEM, n = 10 for each genotype. (J) A schematic diagram of the regulation of Brm activity in intestinal homeostasis. ISCs divide asymmetrically to an EB and an ISC. EBs then differentiate into ECs or ee cells. Cell-type-specific markers are indicated. In normal state (left side), Brm is expressed at a high level in nuclei of ISCs, EBs, and some ee cells, and at a low level in nuclei of ECs. The different Brm protein levels in nuclei are marked by red (ISCs, EBs, and ee cells) or pink (ECs). Decrease of Brm protein level in ISCs reduces the ISC proliferative ability and inhibits EC differentiation (right). See also Figure 2—figure supplement 1.
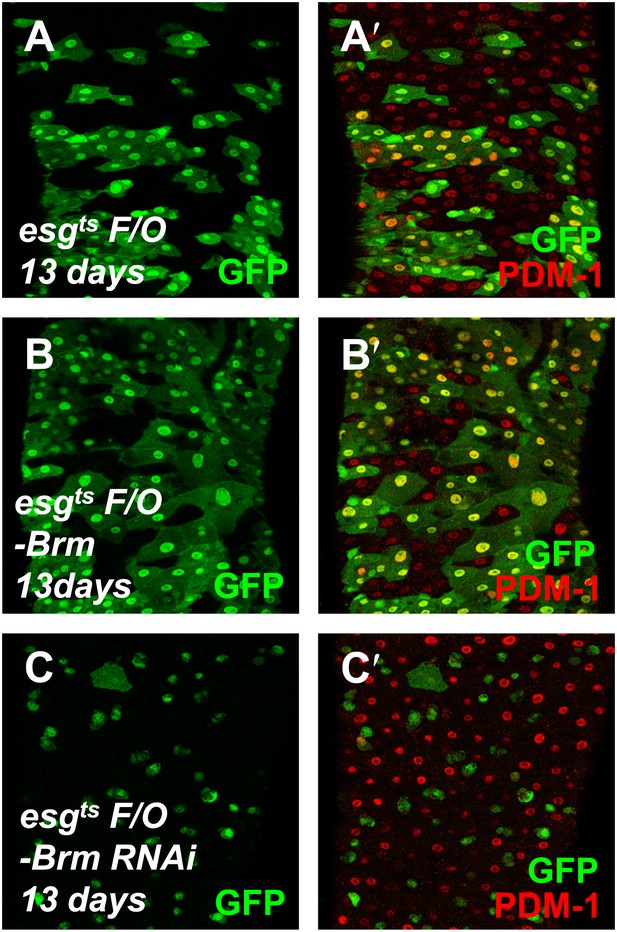
Brm is required for EC differentiation.
(A–C′) The differentiation of adult female midguts was analyzed using the esgts F/O system. Transgenes were induced in the midgut for 13 days. esgts F/O-Brm (B and B′) promoted EC formation, while esgts F/O-Brm RNAi (C and C′) blocked the EC differentiation. PDM-1 marked the EC cells (red).
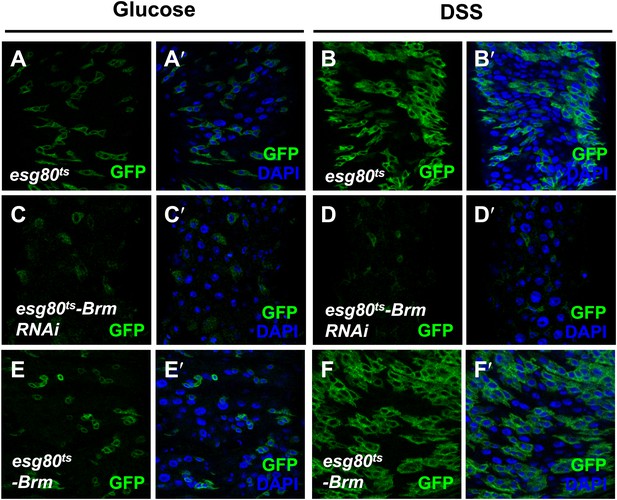
Brm was required for midgut regeneration.
(A–F′) Adult flies expressing esg80ts-Gal4/UAS-GFP (esg80ts) (A–B′), Brm RNAi (esg80ts-Brm RNAi) (C–D′) or esg80ts-Gal4/UAS-GFP-Brm (esg80ts-Brm) (E–F′) were treated with glucose or DSS. Glucose solution with 3% DSS was fed to the flies (B–B′, D–D′, and F–F′) for 3 days before guts dissection.
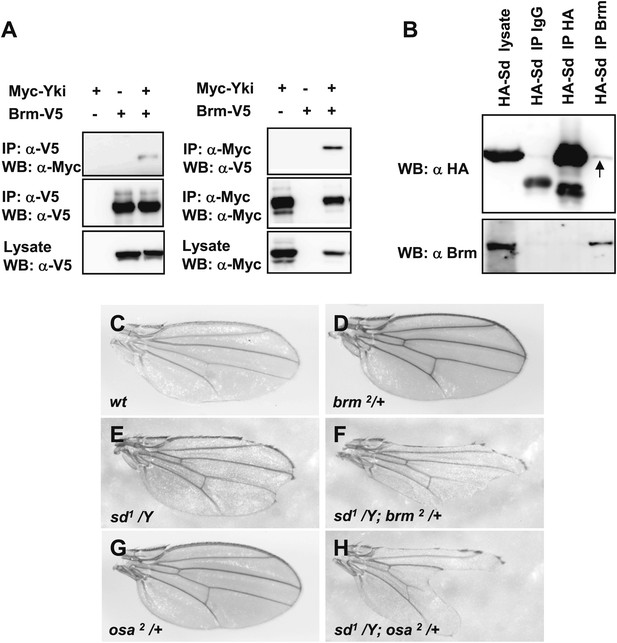
Sd and Yki interact with Brm complex components.
(A) Interaction between overexpressed Myc–Yki and Brm–V5 was detected in S2 cells. Myc–Yki or Brm–V5 was immunoprecipitated with anti-Myc or anti-V5 antibodies. (B) Association between HA–Sd and endogenous Brm in vitro. S2 cells were transfected with the HA–Sd. The arrow indicated HA–Sd coimmunoprecipitated with endogenous Brm. (C–H) Wild-type male wings (C) or hemizygous male wings of null allele brm2/+ (D), or hypomorphic allele sd1/Y (E), or double-mutant combinations of sd1/Y; brm2/+ (F), or hypomorphic allele osa2/+ (G), or combinations of sd1/Y; osa2/+ (H). See also Figure 4—figure supplement 1.
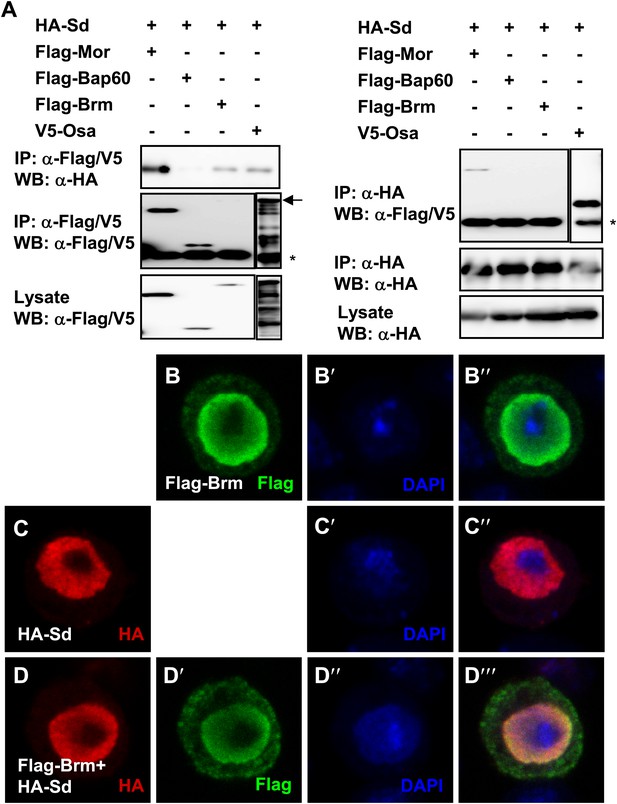
Brm complex associates with Sd.
(A) Sd interacted with Brm, Osa in one direction while in both directions with Mor. The interaction between Sd and Bap60 was not detected. The asterisk marked the band of heavy chain of IgG and the arrow marked the band of Osa. (B–D′′′) Overexpressed Sd and Brm localized in the nuclei of S2 cells. Cells were immunostained with indicated antibodies, HA (red), Flag (green) and DAPI (blue).
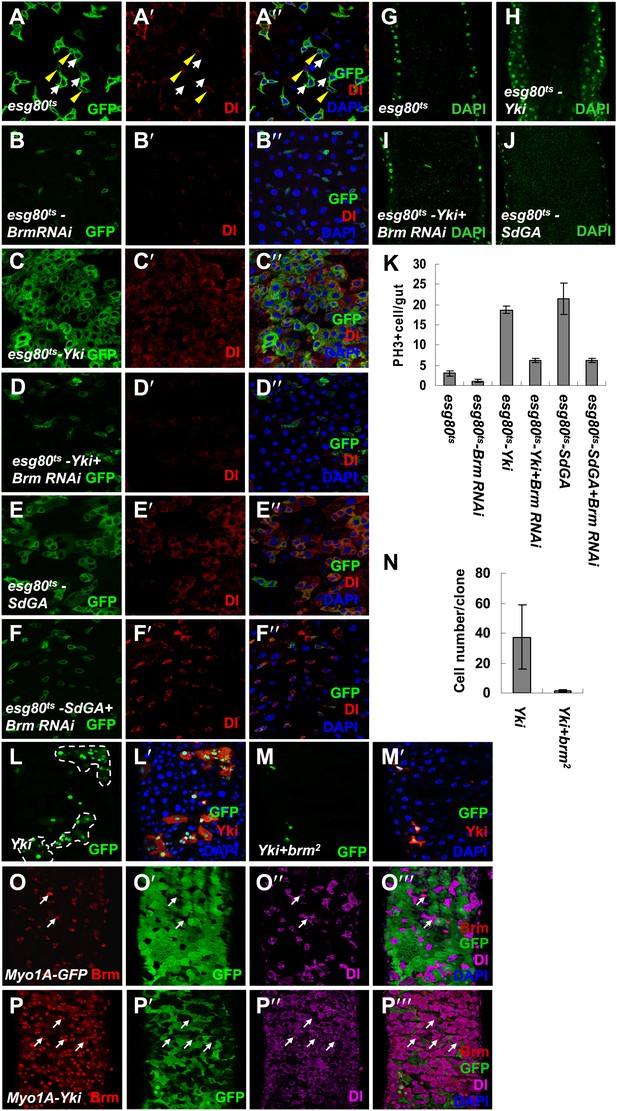
Knockdown of Brm blocks Yki/SdGA-induced ISC proliferation.
(A–F′′) Adult flies expressing esg80ts (A–A′′), esg80ts-Brm RNAi (B–B′′), esg80ts-Yki (C–C′′), esg80ts-Yki+Brm RNAi (D–D′′), esg80ts-SdGA (E–E′′), esg80ts-SdGA +Brm RNAi (F–F′′) were cultured at 29°C for 8–9 days. Midguts were dissected and immunostained for Dl (red) and DAPI (blue). White arrows indicated the EBs, and yellow arrowheads indicated the ISCs. (G–J) Images show an optical cross-section through the center of the intestine, DAPI (green). (K) Quantification of PH3 positive mitotic cells of the indicated guts. The results represent the mean ± SEM, n = 10 for each genotype. (L–M′) Adult midguts containing nuclear localized GFP-labeled control non-tagged form of Yki overexpressed clones (L and L′) or Yki plus brm2 clones (M and M′) were immunostained for Yki (red) and DAPI (blue). Guts were dissected from the adult flies 72 hr after clone induction. (N) Quantification of the cell number of Yki or Yki+brm2 clones. 10 guts were counted for each genotype. (O–P′′′) Adult guts of Myo1A-Gal4 UAS-GFP;tubGal80ts control (O–O′′′) or expressing Myo1A-Gal4 UAS-GFP;tubGal80ts-Yki (P–P′′′) were immunostained for Brm (red), Dl (purple), and DAPI (blue). Arrows indicated ISCs with a high endogenous Brm protein level.
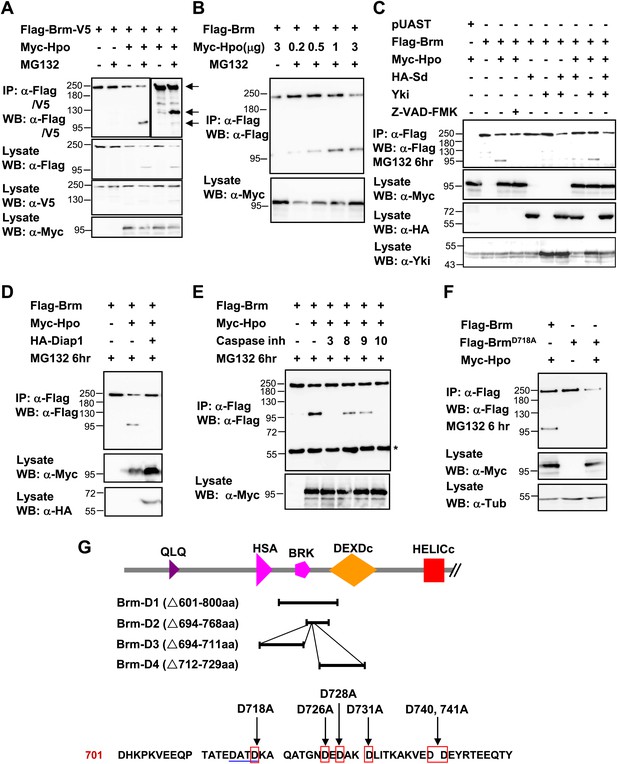
Brm is cleaved at the D718 site by Hpo-induced caspase.
(A) Flag-Brm-V5 was transfected with or without Myc-Hpo. Western blots (anti-Flag or anti-V5) of IP samples were performed to detect the N- or C-terminus of Brm. MG132 was treated 6 hr before harvesting the cells. Arrows indicated the full length Brm (top) and the N- (bottom), C- (middle) terminal cleaved product of Brm. (B) 3 μg of Flag-Brm was cotransfected with different dosages of Hpo plasmids in S2 cells, MG132 was treated 6 hr before harvesting the cells. (C) Cotransfected Flag-Brm and Myc-Hpo together with Sd/Yki or in the presence of caspase inhibitor Z-VAD-FMK, the cleaved Brm fragments were unable to be detected. Z-VAD-FMK was added to a final concentration of 10 mM for 6 hr. (D) S2 cells were transfected with Myc-Hpo and Flag-Brm with HA-Diap1. (E) Flag-Brm and Myc-Hpo were cotransfected in S2 cells to induce the cleavage of Brm. Inhibitors of Caspase 3, 8, 9, 10 were added to block the cleavage in a final concentration of 10 mM for 6 hr. Asterisk indicates IgG bands (loading control). (F) BrmD718A mutation blocked Hpo-induced Brm cleavage in S2 cells. (G) A schematic representation of Brm deletions and mutations. Brm-D1 to D4 were the deletions that were used to map the cleavages site of Brm. Brm-D718A/D726A/D728A/D731A/D740,741A were the mutants generated for mapping the cleavage sites. The novel caspase recognition motif (DATD) in Brm is indicated by a single blue underline including D718 residue. See also Figure 6—figure supplement 1.
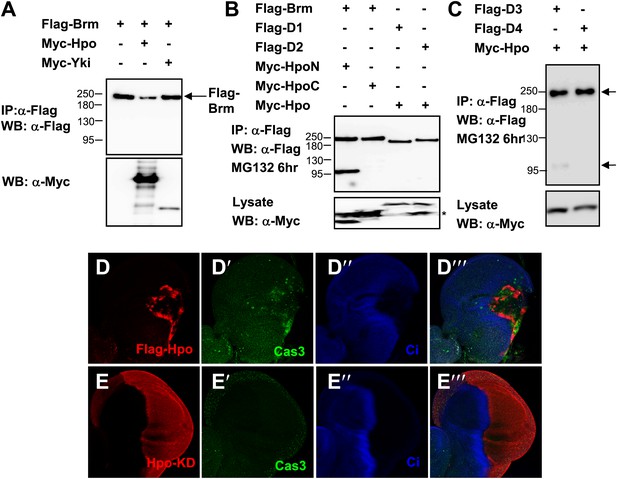
Brm is cleaved by Hpo-induced caspase.
(A) The protein level of Brm was decreased when cotransfected with Myc-Hpo (lane two), but not Myc-Yki (lane three). (B) Hpo N-terminal kinase domain 1–342 aa (Hpo-N) induced Brm cleavage, but not Hpo C-terminal regulatory domain 343 aa-end (Hpo-C) (lane one and lane two). Brm deletions, D1 and D2, failed to produce the 100 KD band when Hpo existed (lane three and lane four). Brm deletions were immunoprecipitated with anti-Flag antibody. (C) Brm deletion D3 still was cleaved when Hpo was cotransfected, but the D4 could not. The full length Brm and the N-terminal cleaved fragment of Brm are marked by arrows. (D–E′′′) Overexpressing Hpo rather than Hpo kinase dead (Hpo-KD) induced the activated caspase 3 signal. Wing discs expressing wild-type UAS- Hpo (D–D′′′) or UAS-Hpo-KD (E–E′′′) under control of hhGal4 driver were immunostained with activated casepase 3 antibody (Cas3, green), Flag antibody (red) and Ci antibody (blue).
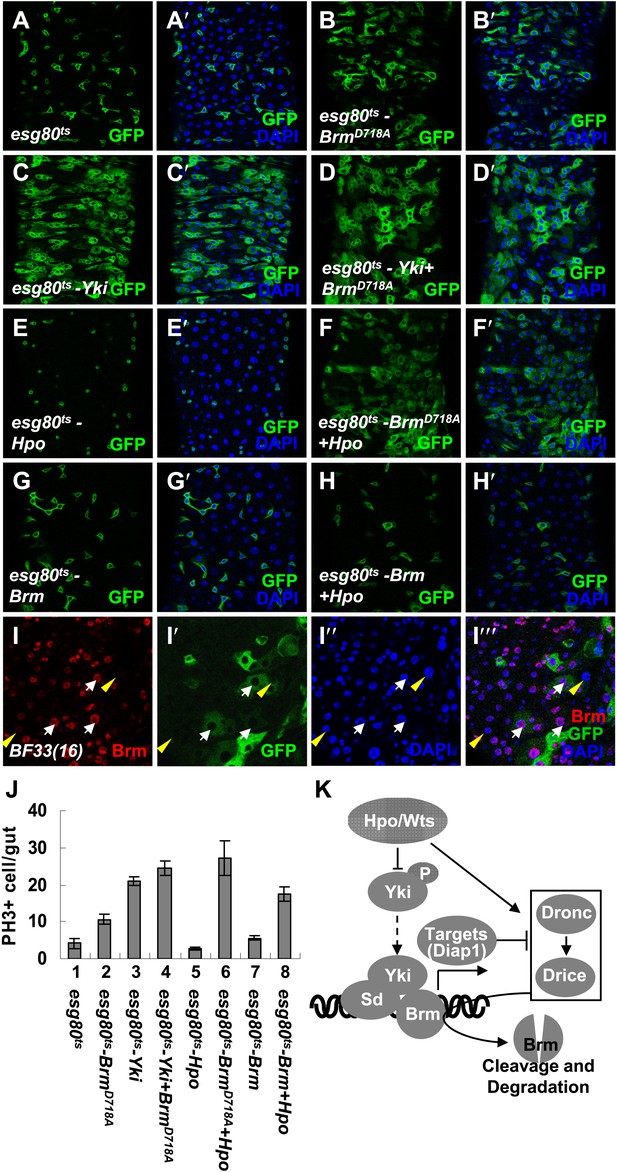
The cleavage resistant mutant BrmD718A promotes ISC proliferation with antagonistic ability against Hpo activity.
(A–H′) Adult guts of esg80ts control (A and A′), esg80ts-BrmD718A (B and B′), esg80ts-Yki (C and C′), esg80ts-Yki+BrmD718A (D and D′), esg80ts-Hpo (E and E′), esg80ts- BrmD718A +Hpo (F and F′), esg80ts-Brm (G and G′) and esg80ts-Brm+Hpo (H and H′) were immunostained for DAPI (blue). (I–I′′′) Adult midguts containing GFP-labeled MARCM clones of hpo null allele (BF33(16)). White arrows indicate the ECs in the BF33(16) clones, and yellow arrowheads indicate the ECs outside the clones. (J) Quantification of PH3 positive mitotic cells of the indicated guts. The results represent the mean ± SEM, n > 10 for each genotype. (K) A model of the regulation of Brm protein stability by the Hpo pathway. The Hpo pathway restricts Brm protein level by inducing the activation of caspase to cleave Brm and/or by inhibiting the expression of Yki–Sd target genes, especially diap1 that inhibits the caspase activity. See also Figure 7—figure supplements 1–4.
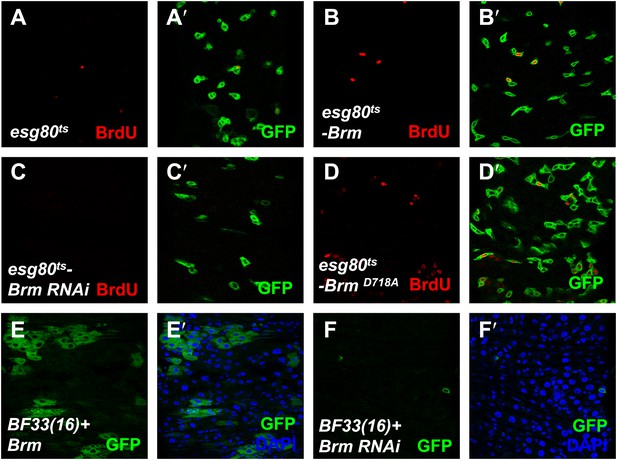
The cleavage resistance mutant BrmD718A promotes ISCs proliferation.
(A–D′) Adult guts of esg80ts-Gal4 control (A and A′), esg80ts-Brm (B and B′), esg80ts-Brm RNAi (C and C′), esg80ts-BrmD718A (D and D′) were immunostained for BrdU (red). Note that BrmD718A increased the BrdU number. (E and F′) Adult midguts containing GFP-labeled MARCM clones of hpo null allele BF33(16)+ Brm (E–E′) or hpo null allele BF33(16)+ Brm RNAi (F–F′). Brm RNAi blocked the cell proliferation in hpo null allele clones.
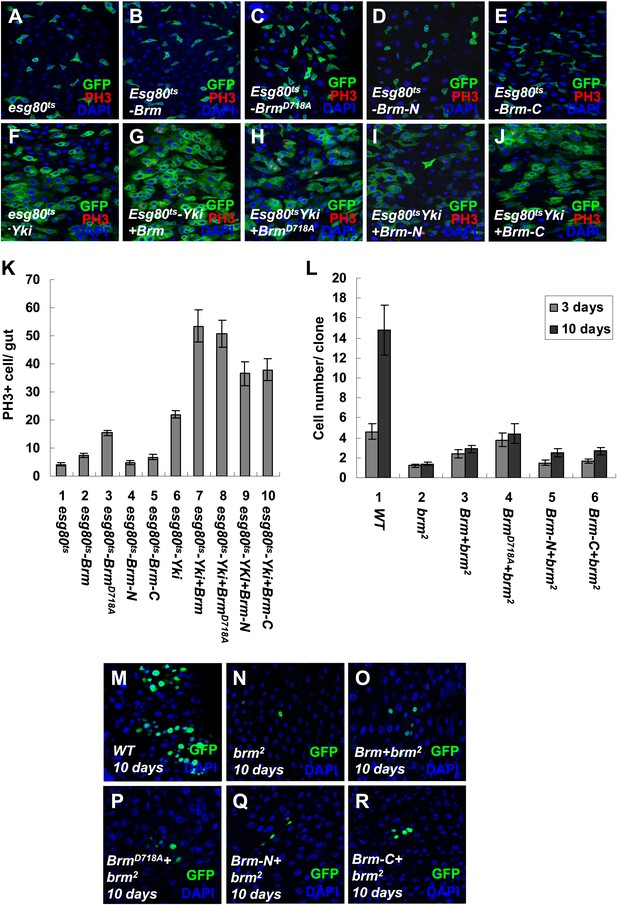
The cleavage products of Brm have low activity in promoting ISC proliferation.
(A–J) Adult guts of esg80ts control (A), esg80ts-Brm (B), esg80ts-BrmD718A (C), esg80ts- esg80ts-Brm-N (D), esg80ts-Brm-C (E), esg80ts-Yki (F), esg80ts-Yki+Brm (G), esg80ts-Yki+BrmD718A (H), esg80ts-Yki+Brm-N (I) and esg80ts-Yki+Brm-C (J) were immunostained for PH3 (red) and DAPI (blue). (K) Quantification of PH3 positive mitotic cells of the indicated guts. The results represent the mean ± SEM, n > 10 for each genotype. (L) Quantification of the cell number of the MARCM clones of the indicated genotypes. Guts were divided into two groups after clone induction: 3 days and 10 days. The results represent the mean ± SEM, n > 10 for each group. (M–R) Adult midguts containing nuclear localized GFP-labeled wild-type control clones (M), brm null allele brm2 clones (N), Brm+brm2 clones (O), BrmD718A+brm2 clones (P), Brm-N+brm2 clones (Q) and Brm-C+brm2 clones (R) were immunostained to show the DAPI (blue). Guts were dissected from the adult flies 10 days after clone induction.
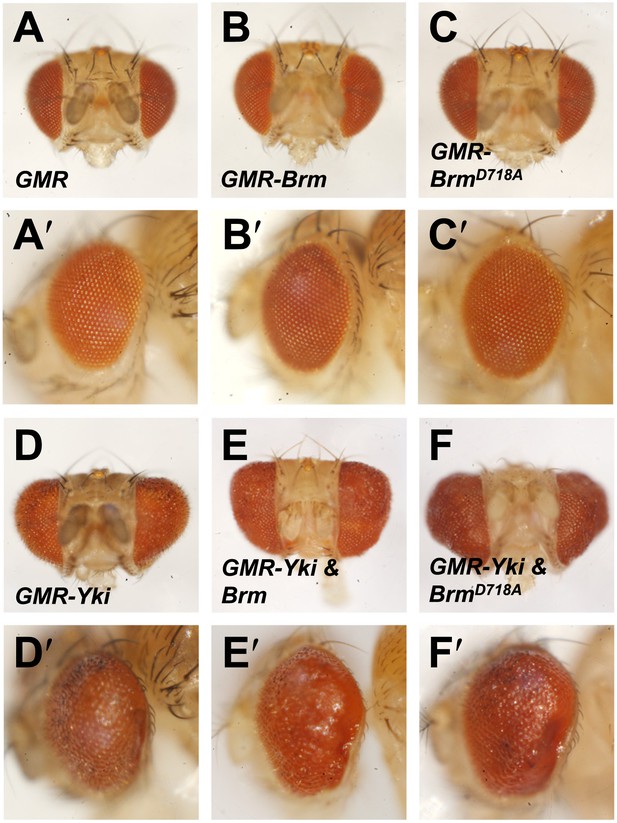
Genetic interaction assays between Brm and Yki/Sd in Drosophila eyes.
(A–F′) Genetic interaction assays between Brm and Yki in Drosophila eyes. BrmD718A further increased Yki-induced eye overgrowth. Adult eyes of GMR (A and A′), GMR-Brm (B and B′), GMR- BrmD718A (C and C′), GMR-Gal4/UAS-Yki (D and D′), GMR-Gal4/UAS-Yki+wild type Brm (E and E′), GMR-Gal4/UAS-Yki+BrmD718A (F and F′).
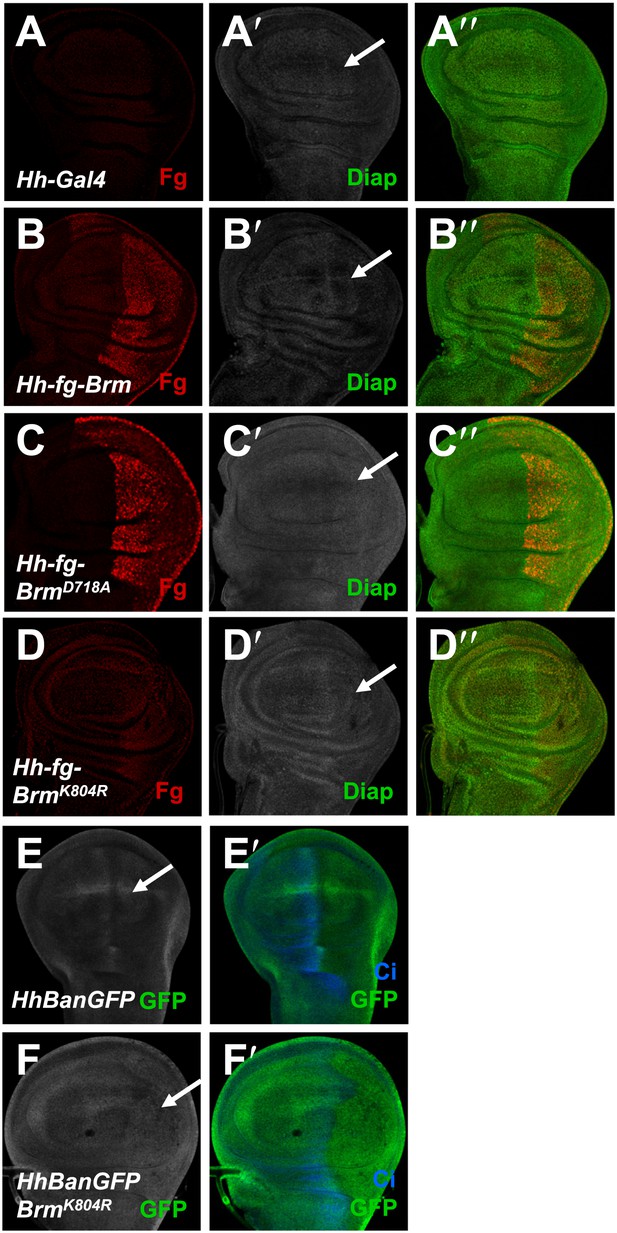
Brm regulates Hpo pathway target genes in wing discs.
(A–D′′) Wing discs of hh-Gal4 control (A–A′′) or expressing UAS-Flag-Brm (B–B′′) or UAS-Flag-BrmD718A (C–C′′) or UAS-Flag-BrmK804R (D–D′′) were immunostained to show the expression of flag (red), and Diap1 (green). P-compartment of the wing discs was marked by arrows. Of note, BrmK804R always shows a weak expression in wing discs. (E–F′) Bantam sensor upregulated in P-compartment of the wing discs when BrmK804R was expressed, which stands for decreased Bantam level in the P-compartment. Wing discs of hhBanGFP control (E and E′) or expressing UAS-Flag-BrmK804R (F and F′) were immunostained to show the expression of Bantam sensor (BanGFP, green) and Ci (blue).
Tables
Mass spectrum analysis results
| Protein description | Molecular function | Pep count | Unique Pep count |
|---|---|---|---|
| Yki mass spectrum | |||
| Brahma (Brm) | ATP-dependent helicase | 11 | 5 |
| Osa | DNA binding | 5 | 4 |
| Sd mass spectrum | |||
| Brahma associated protein 55kD (Bap55) | Structural constituent of cytoskeleton | 8 | 5 |
| Brahma associated protein 60kD (Bap60) | Protein binding | 5 | 3 |
| Brahma (Brm) | ATP-dependent helicase | 4 | 4 |
| Brahma associated protein 155 kDa (Mor) | Protein binding | 1 | 1 |
-
To determine whether there are physical interactions between Yki/Sd transcriptional complex and Brm complex and gain further understanding of the regulation mechanism of Brm in regulating ISC proliferation, we immunoprecipitated endogenous Sd or Yki protein in S2 cells using generated rabbit anti-Sd or anti-Yki antibodies, respectively, followed by mass spectrometry (MS) analysis. The corresponding proteins of Brm complex identified in association with Yki (Yki mass spectrum) or Sd (Sd mass spectrum) are listed with the number of peptides identified by mass spectrometry.





