Crystal structures of virus-like photosystem I complexes from the mesophilic cyanobacterium Synechocystis PCC 6803
Figures
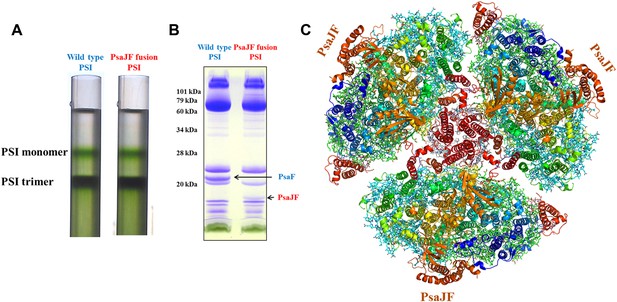
The crystal structure of a PsaJF fusion containing PSI complex.
(A) Sucrose gradients of chlorophyll containing complexes from either wild type or PsaJF containing bacteria showing similar trimer to monomer ratios in both strains. (B) An SDS-PAGE gel showing the polypeptide composition of the trimeric PSI fractions from the sucrose gradient showing the presence of the PsaJF fusion (which was confirmed by MS analysis) in the purified PSI complex. (C) The PSI trimer structure viewed from the stromal side of the membrane showing the fused subunit assembled in the complete complex.
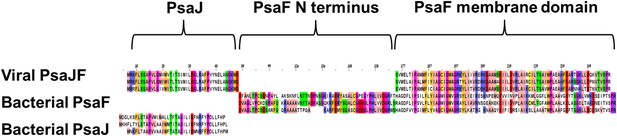
Multiple sequence alignment showing the domain architecture of the viral JF fusion sequences compared to the cyanobacterial PsaF and PsaJ sequences.
https://doi.org/10.7554/eLife.01496.004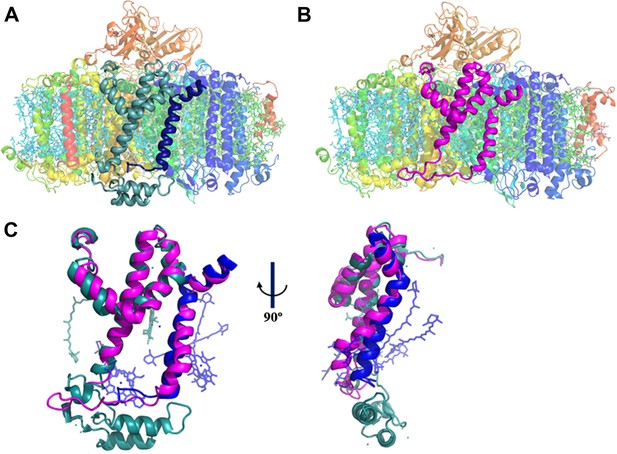
A JF containing PSI complex has a luminal plateau.
(A) PSI from Thermosynechococcus with PsaJ (Blue) and PsaF (cyan) shown from the membrane plane, the N-terminus of PsaF extends as two alpha helices into the luminal space. PsaX is colored red. (B) The PsaJF (magenta) containing PSI complex from Synechocystis showing the flatter luminal plateau created from the removal of the N-terminus of PsaF. (C) Superposition of the JF subunit from Synechocystis (magenta) on the PsaF (cyan) and PsaJ (blue) subunits from Thermosynechococcus, the overall structure of the fused subunit remains undisturbed.
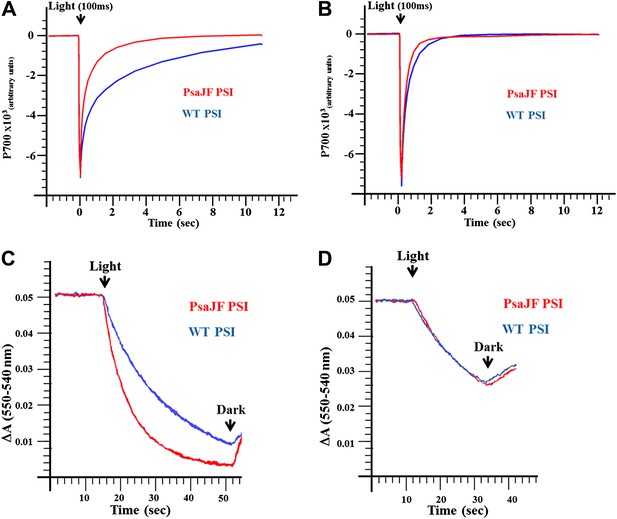
PSIPsaJF is a promiscuous electron acceptor.
(A) P700 reduction kinetics (after a 100 ms pulse of orange light) showing faster reduction kinetics of a PSIPsaJF complex with respiratory CytC (red line) compared to the wild-type complex (blue line). (B) Native cytochrome C553 reduces both wild-type and PSIPsaJF complexes at similar rates. (C) Plots of the cytochrome oxidation (ΔA 550 nm–540 nm) under continuous white illumination by PSI, showing faster oxidation of a respiratory cytochrome by PSIPsaJF (red trace) compared to the wild-type PSI (blue trace). (D) Native CytC553 is oxidized with similar kinetics by both complexes.
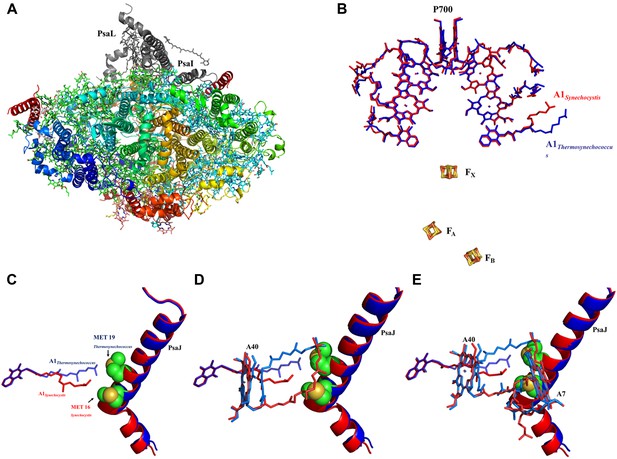
High resolution structure of a monomeric PSI complex from Synechocystis.
(A) A luminal view of the monomeric PSI complex. PsaL and PsaI that are missing from our crystal are shown in gray. (B) Comparison of the ETC of Synechocystis (red) to that of Thermosynechococcus (blue) reveals near perfect superposition in all the components with the clear exception of the quinone A1 isoprenoid tail. (C) Superposed PsaJ from Synechocystis (red) and Thermosynechococcus (blue) showing the coordinating methionine residue (carbons in green and the sulfur atom in yellow). (D) The phytol tail of chlorophyll A40 is reoriented in Synechocystis in order to accommodate the movement of the quinone tail. (E) The phytol tail of Chlorophyll A1 in Synechocystis shifted to accommodate the PsaJ MET16 change.
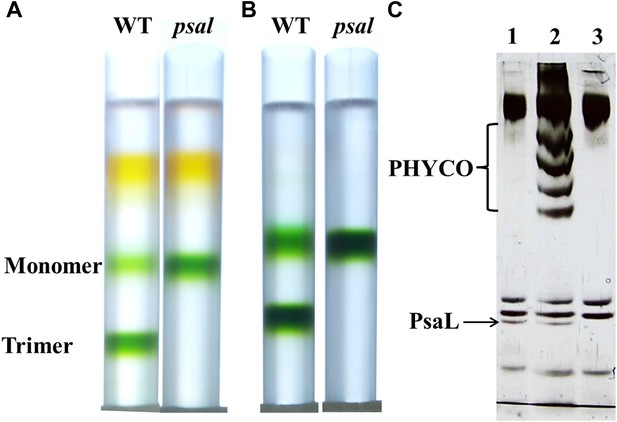
Purification of a monomeric PSI from a Synechocystis psal mutant.
(A) Sucrose gradients of dissolved membranes from wild-type or psal strains showing a homogenous population of monomeric complex in the psal mutant. (B) Sucrose gradients of DEAE chlorophyll peak fractions from either wild-type or psal strains. (C) Silver stained SDS-PAGE of the polypeptide composition of the chlorophyll peaks (0.5 µg chlorophyll loaded in each well) from B (lane 1–WT trimer, lane 2–WT monomer, lane 3–psal monomer) showing the absence of PsaL in the psal mutant. The four bands that co-purified with the monomeric PSI from the wild-type sample (labeled ‘PHYCO’) were shown by MS analysis to contain CP47, CP43, Lcm and isiA, these bands were consistently absent from the psal mutant complex preparations.
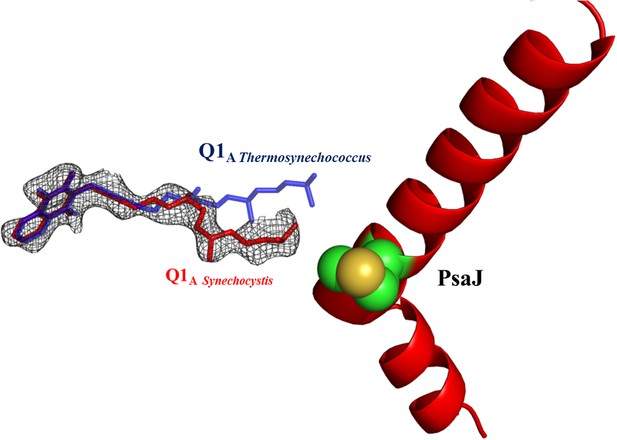
Electron density map of Q1A showing the different position of the A1 quinone from Synechocystis (in red).
2Fo-Fc maps were contoured at 1.4σ.
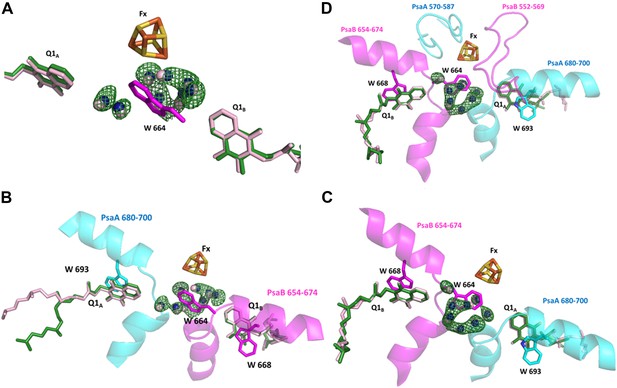
Conservation of a water pocket lining the electron path between Q1B and Fx.
(A) Electron transfer (ET) pathways between Q1A, Q1B and Fx. Positive difference map peaks calculated with the water molecules omitted from the model are shown in green (contoured at 3σ). Quinones and waters from Thermosynechococcus are colored in light pink. Synechocystis quinones are colored in green, water in blue, and amino acids in magenta. (B) Same as A only the relevant fragments from PsaA and PsaB from Synechocystis are included. Quinone coordinating Trp residues are shown (W 693 and W 668 from PsaA and PsaB respectively), together with Trp 664 in PsaB which separates Fx and Q1A (The corresponding amino acid in PsaA is Glycine 689). (C) A 180° rotation of B showing a different view of the water pocket. (D) A view of the entire Fx binding regions shown with the Q1 and Fx coordination loops.

Multiple sequence alignment (MSA) of PsaJ sequences from various cyanobacteria.
Position 16 and 19 in the Synechocystis PsaJ sequence are indicated on top. Sequences were aligned using muscle and rendered with Jalview8.
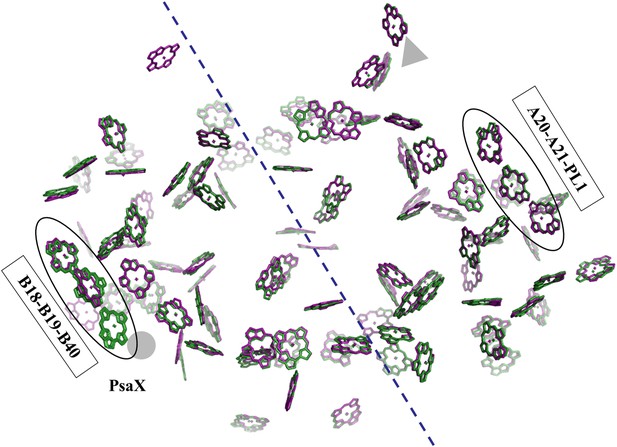
Pseudo C2 symmetry in the antenna system of Synechocystis.
A depth cued image comparing the overall organization of the PSI antenna from Synechocystis (green) or Thermosynechococcus (purple) viewed from the stromal side of the membrane, only chlorine rings are shown for clarity. The C2 pseudosymmetry axis is shown in blue. PsaX is shown as gray filled circle. The new chlorophyll detected in Synechocystis (B40) is clearly seen next to PsaX which precludes its binding in Thermosynechococcus.
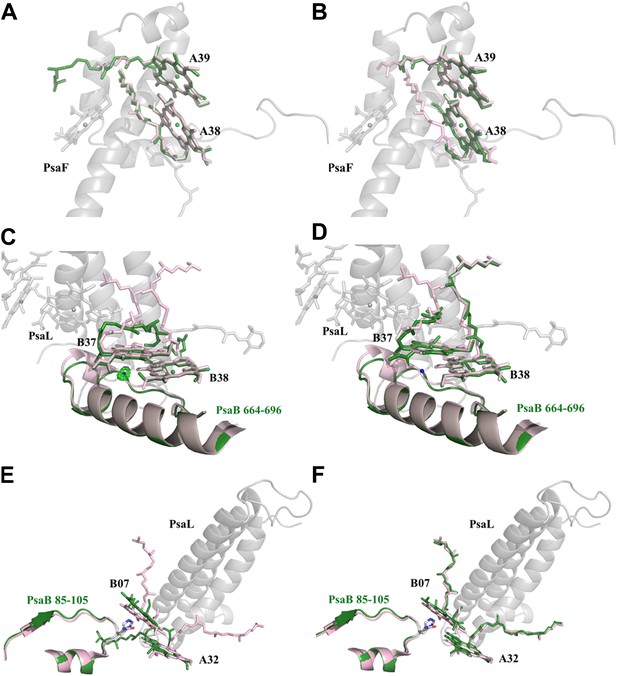
Red chlorophylls conformations in Synechocystis monomeric and trimer PSI.
(A) Synechocystis monomeric conformation (green) of chlorophyll pair A38-A39 compared to Thermosynechococcus conformation in light pink. (B) The conformation of the A38-A39 pair in the PSIPsaJF trimeric complex shows no significant movements between all three PSI's complexes. PsaF (from the monomeric complex) is shown in the background in gray. (C) Conformation of the putative red pair B37-B38 as observed in our monomeric model. The coordinating water molecule of B37 is shown as a blue sphere (the position of the corresponding water from Thermosynechococcus is in pink) together with the Fo-Fc omit map (contoured at 4σ). The Large movements of the phytol tails probably result from the absence of PsaL since they are absent in the trimer PSIPsaJF model, shown in (D). (E) Synechocystis monomeric conformation (green) of chlorophyll pair B07-A32. The significant movement of the chlorine ring of B07 is clearly seen. The three transmembrane helixes of PsaL missing in the Synechocystis monomeric crystal are shown together with the conformation of this region in Thermosynechococcus (light pink). (F) The same region as modeled in the 3.8 Å PSIPsaJF trimer (Synechocystis in green, Thermosynechococcus in light pink) does not show any movements, however the low resolution of the trimetric model prevents us from reaching a definite conclusion.
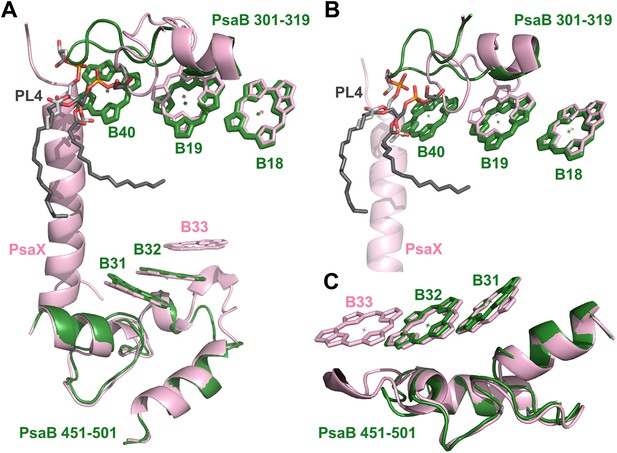
Complementing structural rearrangements on opposite sides of PSI in Synechocystis.
(A) The luminal-side chlorophyll trimer (B31-B32-B33) is missing in Synechocystis, while chlorophyll B40 forms a stromal chlorophyll trimer (Synechocystis in green, Thermosynechococcus in light pink). The PsaB loop on the stromal side (PsaB 301-319) extends to coordinate chlorophyll B40 and A phospholipid (shown in dark gray), while the luminal side PsaB loop (PsaB 451-501) is shortened in Synechocystis and chlorophyll B33 is lost as a result. (B) A close up on the stromal side changes showing the large conformational change in PsaB. In spite of this the phospholipid is clearly observed. (C) Loss of the B31-B33-B33 chlorophyll trimer in Synechocystis, the shortened PsaB loop together with the remaining chlorophylls in Synechocystis is shown.
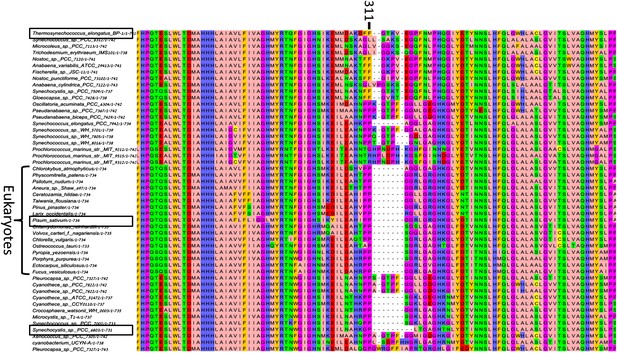
High variability in the B31-B32-B33 supporting loop of PsaB.
Representative sequences (50 out of 200 used for alignment) from MSA of various PsaB sequences, position 311 in the Synechocystis PsaB sequence is indicated on top. Boxes highlight sequences from the three known PSI structures.
Tables
X-ray data collection and refinement statistics
| Data collection | PsaJF trimer | PSI monomer | |
|---|---|---|---|
| Beamline | ESRF–ID29 | ESRF–ID29 | SLS–PXI–X06SA |
| Wavelength (Å) | 0.97625 | 0.97625 | 1 |
| Resolution (Å) | 30–3.8 | 30–2.8 | 30–3 |
| Measured reflections | 426,209 (63,882) | 419,672 (61,558) | 396,647 (57,225) |
| Unique reflections | 113,221 (16,536) | 91,895 (13,209) | 72,095 (10,585) |
| Rpim (%) | 6.8 (71) | 7.5 (124) | 5.6 (77.3) |
| I/σ(I) | 9.9 (1.2) | 8 (1.3) | 10 (1.2) |
| Completeness | 98.9 (99.5) | 99.4 (99.1) | 96.8 (98.3) |
| Redundancy | 3.8 (3.9) | 4.6 (4.7) | 5.1 (5.4) |
| Space group | P 21 | P 21 21 21 | P 21 21 21 |
| Unit cell dimensions | |||
| a, b, c (Å) | 214, 134, 220 | 120, 173, 179 | 120, 174, 179 |
| α, β, γ (°) | 90, 111.1, 90 | 90, 90, 90 | 90, 90, 90 |
| Refinement statistics | |||
| Resolution (Å) | 30–3.8 | 30–2.8 | 30–3 |
| Rwork/Rfree | 25.9/29.7 | 21/24 | 24.4/28 |
| No. of chains | 30 | 9 | 9 |
| No. of ligands | 360 | 119 | 116 |
| Average B-factor (Å2) | 128 | 85 | 90 |
| R.M.S deviations | |||
| Bond Angles | 1.7 | 1.7 | 1.7 |
| Bond lengths | 0.014 | 0.005 | 0.004 |
| Ramachandran statistics | |||
| Favored region % | 93.8 | 93.8 | 93.2 |
| Allowed region % | 5.1 | 5.9 | 6.3 |
| Outlier region % | 1.1 | 0.3 | 0.5 |
| clashscore | 5.5 | 3.8 | 4.6 |
Additional files
-
Supplementary file 1
List of primers used in the construction of PsaJF. PsaJ was amplified with primers 5580 and 5581 and fused to PsaF fragment amplified with primers 5582 and 5583. The JF fusion gene was than ampified using ∼300 bp sequences containing the PsaF promoter and the PsaF down homology to create the entire gene cassette. This 970 bp fragment was cloned into pGEM-Teasy. Finally, the fragment was moved into a pET28 vector in order to introduce the Kan resistance gene into a PstI site that was designed into primer 5584.
- https://doi.org/10.7554/eLife.01496.017





