p97-dependent retrotranslocation and proteolytic processing govern formation of active Nrf1 upon proteasome inhibition
Figures
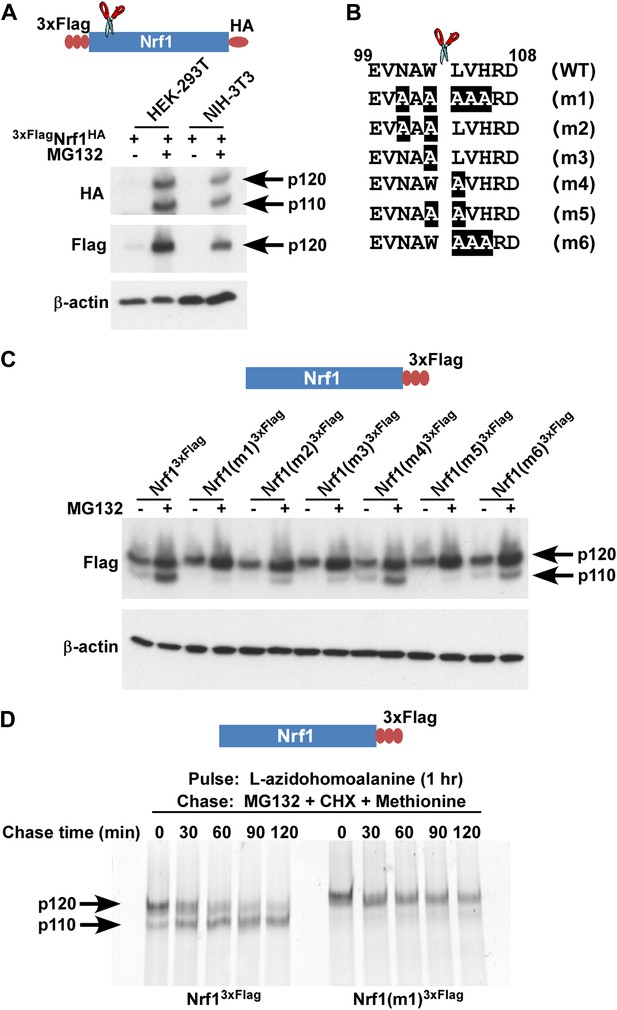
Nrf1 p110 is derived from p120 by proteolytic processing.
(A) Human HEK-293T or mouse NIH-3T3 cells were transduced with a retrovirus expressing 3xFlagNrf1HA and 72 hr later were left untreated or treated with 5 µM MG132 for 5 hr. The cell lysates were then used for immunoblotting with anti-Flag and anti-HA antibodies. β-actin protein levels were evaluated as a loading control. (B) Amino acids 99 to 108 from human Nrf1 are shown. The scissor indicates the cleavage position predicted by the Edman degradation sequencing. Also shown are mutants 1 through 6 with the mutated amino acids highlighted. (C) HEK-293T cells were transiently transfected with wild-type or various mutant constructs (m1 through m6) of Nrf13×Flag and 48 hr later were left untreated or treated with 5 µM MG132 for 5 hr. The cell lysates were then processed for immunoblotting with anti-Flag antibody. β-actin was used as a loading control. (D) HEK-293 cells stably expressing either wild-type Nrf13×Flag or Nrf1(m1)3×Flag were pulse-labeled for 1 hr with L-azidohomoalanine and then chased with MG132, cycloheximide (CHX), and excess methionine. The cells were harvested at the time points indicated (from 0 min to 120 min) and the lysates were subjected to immunoprecipitation with anti-Flag beads. Nrf1 species were visualized using the tetramethylrhodamine based click-chemistry method as described in ‘Materials and methods’.
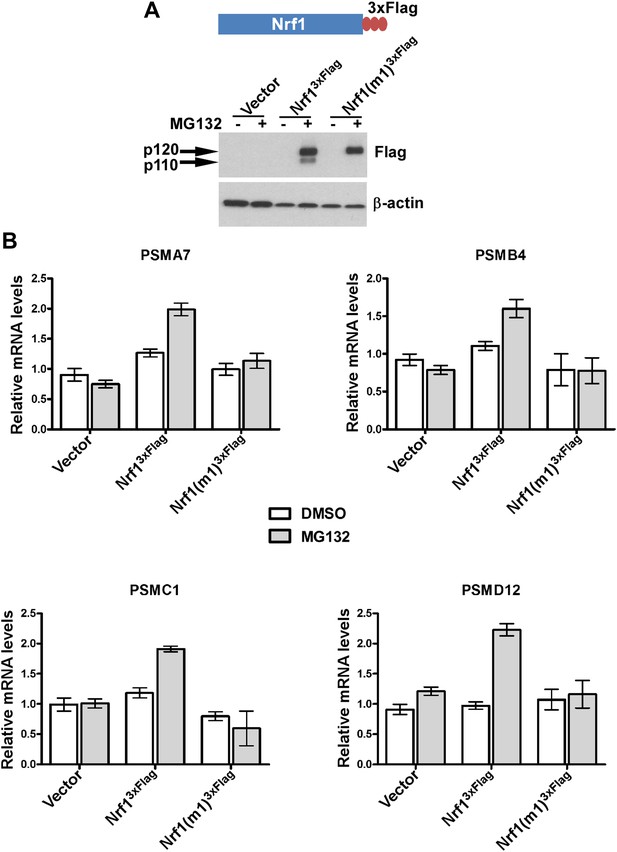
Non-cleavable Nrf1 mutant is functionally defective.
(A) Nrf1−/− MEFs were transfected with constructs expressing either Nrf13×Flag, Nrf1(m1)3×Flag, or vector control and 48 hr later were subjected to Geneticin selection. These antibiotic resistant cells were treated with 1 µM MG132 for 10 hr as indicated. Lysates from these cells were immunoblotted with anti-Flag antibodies to detect exogenous Nrf1. β-actin was used as a loading control. (B) RNA from Nrf1−/− MEFs transfected and selected as described above was used for quantitative RT-PCR to assess the mRNA levels of representative PSM genes. The values were normalized to GAPDH mRNA levels. Error bars denote SD (n = 3).
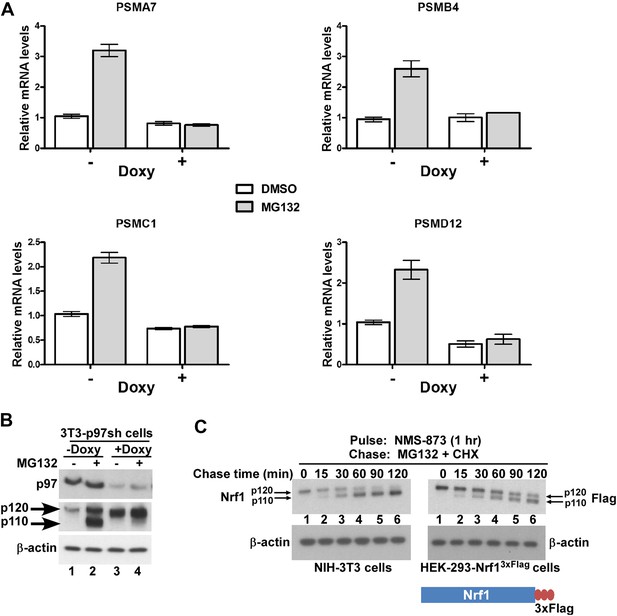
p97 is required for processing and activation of Nrf1.
(A) NIH-3T3-p97sh cells stably expressing doxycycline (Doxy)-inducible shRNA targeting p97 were either mock-treated or induced with Doxy (1 µg/ml) for 3 days after which the cells were further treated with DMSO or 1 µM MG132 as indicated for 10 hr. The RNA from these cells was used for quantitative RT-PCR to assess the mRNA levels of the representative PSM genes. The values were normalized to GAPDH mRNA levels. Error bars denote SD (n = 3). (B) NIH-3T3-p97sh cells were subjected to Doxy and MG132 treatments as above and the cell lysates were fractionated by SDS-PAGE and immunoblotted to detect p97 and Nrf1. β-actin was used as a loading control. (C) NIH-3T3 (left panel) or HEK-293-Nrf13×Flag cells (right panel) were pulsed for an hour with 10 µM NMS-873 and then chased with MG132 plus cycloheximide (CHX). The cells were harvested at the time points indicated (from 0 min to 120 min) and the lysates were fractionated by SDS-PAGE and immunoblotted to detect endogenous Nrf1. β-actin was used as a loading control.
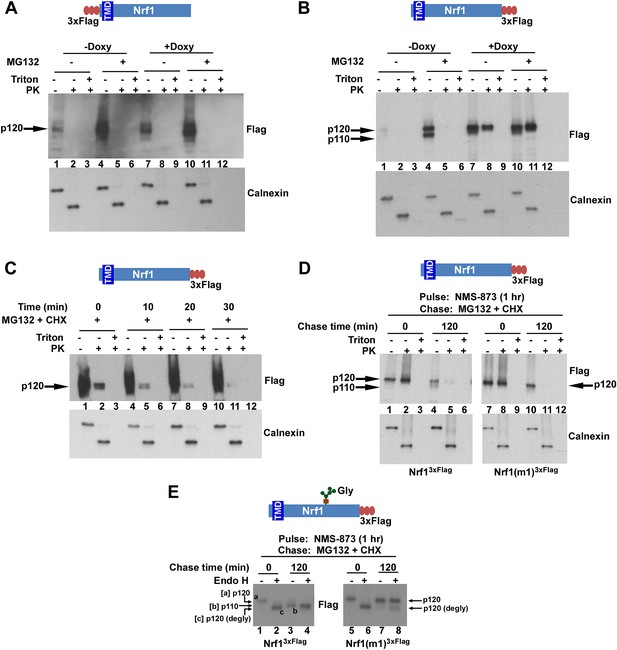
The C-terminus of Nrf1 is re-positioned from the lumen to the cytosol in a p97-dependent manner.
(A) HEK-293 cells stably expressing doxycycline (Doxy)-inducible shRNA targeting p97 (HEK-293-p97sh cells) were transiently transfected with a 3×FlagNrf1 expression construct and 24 hr later were incubated in the presence or absence of Doxy for 4 days after which the cells were further treated or not with 5 µM MG132 for 5 hr. Microsomes prepared from these cells were subjected to protease protection assay with Proteinase K (PK). SDS-PAGE followed by immunoblotting with anti-Flag antibody was used to detect Nrf1. Formation of a discrete cleavage product of Calnexin in the absence of detergent (Triton X-100) was used as an indicator for intact microsomes. (B) Same as (A), except that cells were transfected with an Nrf13×Flag expression construct. (C) HEK-293 cells stably expressing the non-cleavable Nrf1(m1)3×Flag were treated with MG132 plus cycloheximide (CHX) and cells were harvested at indicated time points. Microsomes from these cells were subjected to protease protection assay with PK. Samples were fractionated by SDS-PAGE followed by immunoblotting with anti-Flag antibody. (D) HEK-293 cells stably expressing either Nrf13×Flag or the non-cleavable Nrf1(m1)3×Flag were pulsed for an hour with 10 µM NMS-873 and then chased with MG132 plus cycloheximide (CHX). Microsomes prepared from cells harvested at the indicated time points (0 min and 120 min) were used in a protease protection assay with PK. SDS-PAGE followed by immunoblotting with anti-Flag antibody was used to detect Nrf1. (E) Total cell lysates from the pulse-chase experiment described above were subjected to deglycosylation by the enzyme Endoglycosidase H prior to SDS-PAGE and immunoblotting with anti-Flag. Species ‘a’ refers to full-length Nrf1 (p120) that was fully glycosylated. Species ‘b’ refers to Nrf1 that was proteolytically processed to p110 and degylcosylated following p97-dependent retrotranslocation. Species ‘c’ refers to species ‘a’ that was deglycosylated with Endo H. Note that species ‘b’ migrates slightly more slowly than species ‘c’ even though both species were deglycosylated and species ‘b’ lacked the N-terminal 103 residues. The unexpectedly slow mobility of species ‘b’ presumably reflects acquisition of additional post-translational modifications upon retrotranslocation on Nrf1’s CTD to the cytosolic side of the ER membrane. (TMD–transmembrane domain; Gly–N-linked glycan).
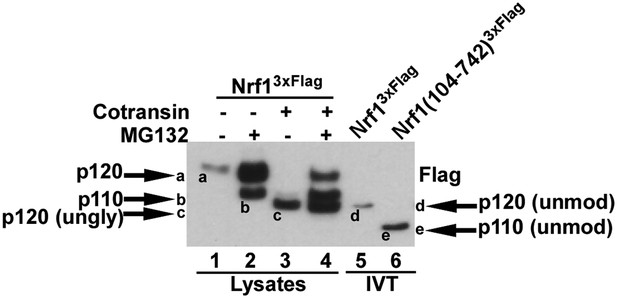
Different forms of Nrf1.
HEK-293 cells stably expressing wild-type Nrf13×Flag were treated with MG132 and/or cotransin, an inhibitor of protein insertion into the Sec61 translocation channel (Garrison et al., 2005) as indicated (lanes 1 to 4) and total cell lysates were prepared. Lanes 5 and 6 contain full-length Nrf13×Flag and Nrf1(104-742)3×Flag that were translated in vitro (IVT) in rabbit reticulocyte lysate in the absence of membranes. The HEK293 cell lysates and IVT reactions were examined by SDS-PAGE followed by immunoblotting with anti-Flag antibody. Different Nrf1 species are shown (ungly–unglycosylated; unmod–unmodified). Nrf1 p120 (species ‘a’) was converted to species ‘c’, which comigrated with the primary translation product (species ‘d’) upon expression in cells treated with cotransin, suggesting that the slow mobility of p120 arose from modifications (e.g., N-linked glycosylation) that occurred within the endoplasmic reticulum. Retrotranslocation and processing of p120 (species ‘a’) yielded p110 (species ‘b’). Species ‘b’ was not sensitive to endoglycosidase H (Figure 4E), suggesting that it was deglycosylated upon retrotranslocation into the cytosol. Nevertheless, species ‘b’ migrated considerably more slowly than the primary translation product for Nrf1(104-742)3×Flag (species ‘e’), indicating that p110 must carry additional modifications that remain uncharacterized. Please note that deglycosylation by cytosolic enzymes converts the Asn at the site of glycosylation to Asp, which could influence migration on SDS-PAGE.
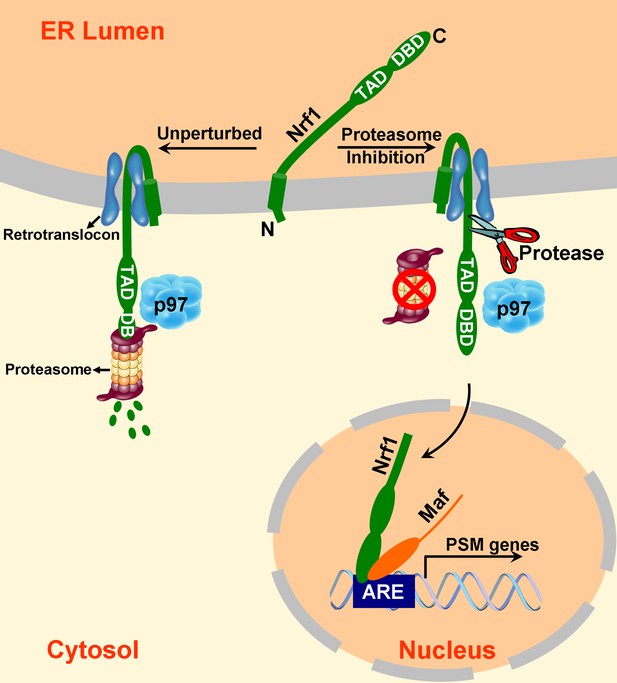
A model to explain Nrf1 activation.
Newly synthesized Nrf1 p120 is initially inserted into the endoplasmic reticulum (ER) membrane in an Ncytosol/Clumen orientation. It is then rapidly and efficiently retrotranslocated to the cytosol, where it is immediately degraded by the proteasome. In cells deficient in proteasome activity, degradation of cytosolically-exposed Nrf1 is retarded, allowing sufficient time for proteolytic processing of Nrf1 p120 to yield the active p110 form which can then migrate to the nucleus, heterodimerize with small Maf proteins, and activate transcription of its targets (e.g., PSM genes) by binding to antioxidant response elements (ARE). Note that the exact mechanism of the retrotranslocation step is unknown; what is shown in the figure reflects one possibility among several. Also, although the putative protease is depicted to be in the cytosol, it remains possible that it is in the ER membrane or lumen.





