FBXO24 modulates mRNA alternative splicing and MIWI degradation and is required for normal sperm formation and male fertility
Figures
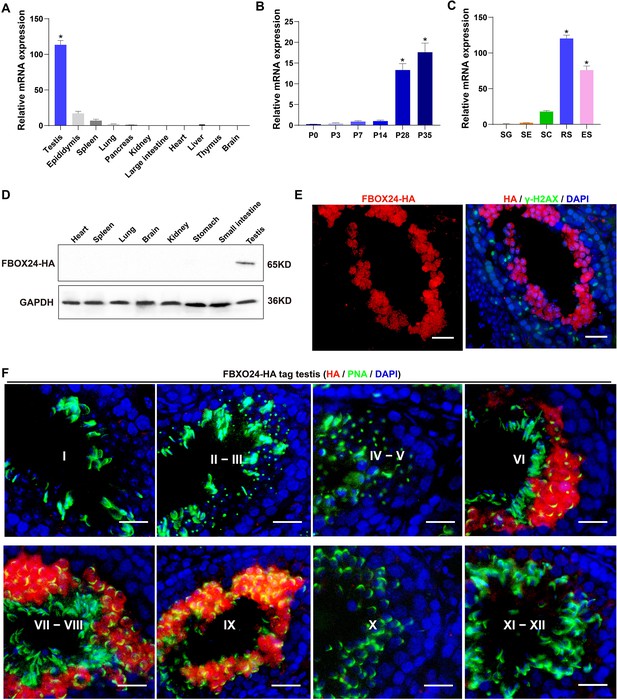
Expression profiles of FBXO24 during testicular development and spermatogenesis in mice.
(A) qPCR analysis of FBXO24 mRNA levels in multiple organs in mice. n = 3/group. Data are mean ± standard deviation (SD). *p < 0.05. (B) qPCR analysis of FBXO24 mRNA levels in developing testes at postnatal day 0 (P0), P3, P7, P14, P28, and P35. n = 3/group. Data are mean ± SD. *p < 0.05. (C) qPCR analysis of FBXO24 mRNA levels in isolated spermatogenic cell populations, including spermatogonia (SG), Sertoli cells (SE), spermatocytes (SC), round spermatids (RS), and elongating spermatids (ES). n = 3/group. Data are mean ± SD. *p < 0.05. (D) Western blot analysis of FBXO24-HA expression in the tissues of adult transgenic mice. (E–F) Co-immunostaining of FBXO24-HA red) and (E) γH2AX (green) or (F) PNA (green) in adult Fbxo24-HA testes. Scale bars = 25 μm. DNA (blue) is stained by DAPI (4,'6-Diamidin-2-phenylindole dihydrochloride). Spermatogenic stages are noted.
-
Figure 1—source data 1
Raw western blot for Figure 1D.
- https://cdn.elifesciences.org/articles/91666/elife-91666-fig1-data1-v1.zip

Expression profiles of FBXO24 in mice and human.
(A) A high degree of conservation of FBXO24 in amino acid sequences among nine species. (B) Amino acid sequence similarity of F-box and regulator of chromatin condensation (RCC1) of FBXO24 protein in mouse and human. (C) mRNA levels of FBXO24 in multiple human organs from GE-mini database. (D) Schematic representation of transgenic cassette.
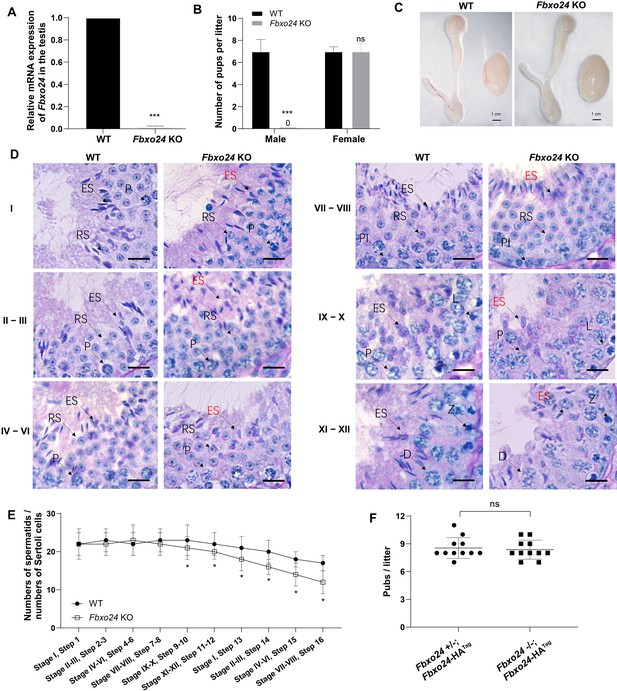
FBXO24 deletion impairs spermatogenic defects in the late steps of spermiogenesis.
(A) qPCR analysis indicates that FBXO24 mRNA is markedly decreased in the testis of Fbxo24 knockout (KO) as compared to wild-type (WT). n = 3 mice /group. ***p < 0.001. (B) The fertility tests of WT and Fbxo24 KO male mice mated with fertile female mice are shown. n = 3 mice /group. Error bars represent mean ± standard deviation (SD). ***p < 0.001; ns: not significant. (C) Histological images of testes and epididymis of WT and Fbxo24 KO mice at 8 weeks old are shown. (D) Periodic acid-Schiff (PAS)–hematoxylin staining of Fbxo24 KO testis at 8 weeks old contained less- condensed late spermatids (red arrows). Spermatogenic stages are noted. RS, round spermatids; ES, elongating spermatids; Pl, preleptotene; P, pachytene; Z, zygotene; D, diplotene. Scale bars = 25 μm. (E) The number of late spermatids is significantly reduced in Fbxo24 KO testis. Ratios of spermatids and Sertoli cells in tubule cross-sections of specific stages of seminiferous epithelial cycles and corresponding spermatid development steps are shown. n = 3 mice. Data are mean ± SD. *p < 0.05. (F) Litter sizes of mating tests. F1 generation of intercrosses between the indicated males and Fbxo24+/− females are shown. Each dot represents one litter.
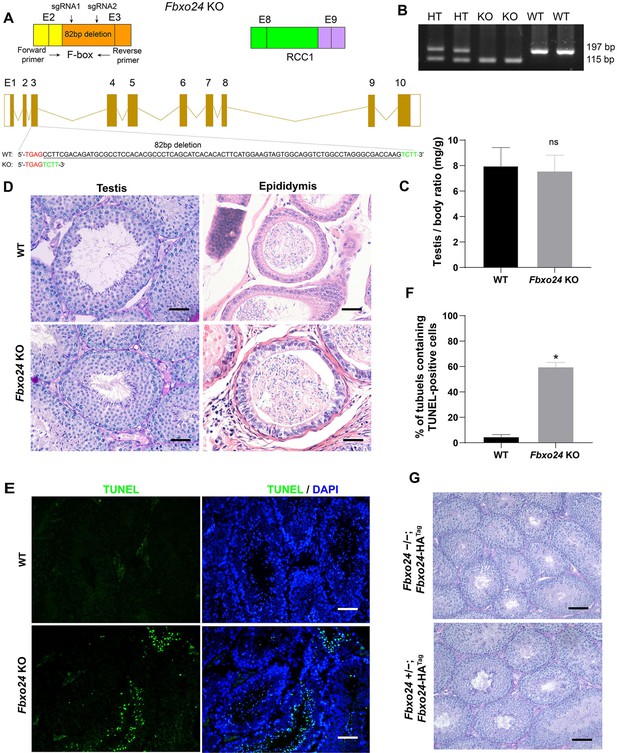
Spermiogenesis was defective in FBXO24-deficient mice.
(A) Diagram illustrating the CRISPR/Cas9 targeting strategy, including position and sequence of guide RNAs (sgRNAs). E, exon. RCC1, regulator of chromosome condensation. (B) Examples of PCR genotyping of the FBXO24 mutated region in WT (wild-type), HT (heterozygote), and KO (knockout) mice. (C) Testis/body weight ratio of WT and Fbxo24 KO (n = 3/group) mice at 8-week-old. Data are mean ± standard deviation (SD). ns, not significant. (D) Representative histological section images of testis and epididymis obtained from Fbxo24 KO mice and WT mice stained with periodic acid-Schiff (PAS) and hematoxylin and eosin (H&E), respectively. Scale bars = 50 mm. (E) TUNEL analysis of WT and Fbxo24 KO testis are shown. Apoptotic cells were labeled by TUNEL staining (green). Scale bars = 50 μm. (F) Comparison of TUNEL-positive seminiferous tubules in WT and Fbxo24 KO testis (n = 3/group). Error bars represent mean ± SD. *p < 0.05. (G) PAS staining for testicular sections of 8-week-old Fbxo24−/−; Fbxo24-HATag mice. Scale bars = 50 μm.
-
Figure 2—figure supplement 1—source data 1
Raw RT-PCR (Reverse transcriptase PCR) gel for Figure 2—figure supplement 1B.
- https://cdn.elifesciences.org/articles/91666/elife-91666-fig2-figsupp1-data1-v1.zip
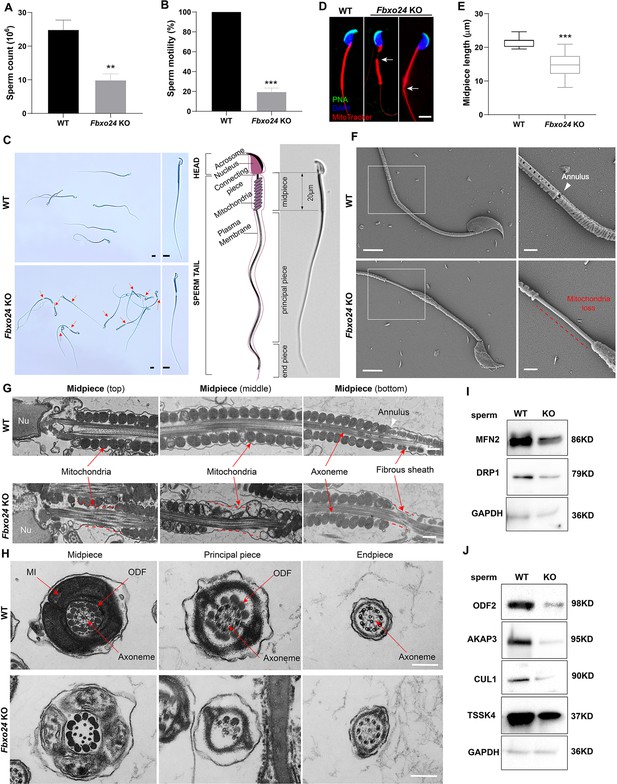
Sperm mitochondria and flagella are defective in FBXO24-deficient mice.
Quantification of sperm counts (A) and sperm motility (B) from wild-type (WT) and Fbxo24 knockout (KO) epididymis are shown. n = 3/group. Error bars represent mean ± standard deviation (SD). **p < 0.01. ***p < 0.001. (C) Sperm morphological images show the defective sperm of Fbxo24 KO mice. Red arrows indicate abnormal gaps in the mitochondrial sheath. Scale bars = 5 μm. (D) Immunofluorescence images of sperm from WT and Fbxo24 KO epididymis. PNA (acrosome, green), MitoTracker (mitochondria, red), and DAPI (nucleus, blue). White arrows indicate the weak or absent staining of MitoTracker. Scale bars = 5 μm. (E) Quantifications of the length of sperm midpiece from WT and Fbxo24 KO mice are shown. n = 100/group. Error bars represent mean ± SD. ***p < 0.001. (F) Scanning electronic microscopy (SEM) images indicate the mitochondria detachment from the flagellum of Fbxo24 KO sperm. Right panel insets show higher magnification of sperm midpiece. The arrowheads indicate the annulus. The dashed red line indicates a region where mitochondria are absent. Scale bars = 2 μm. (G) Transmission electronic microscopy (TEM) images indicate the mitochondria defects in three regions of Fbxo24 KO sperm midpiece in the longitudinal sections. Nu, nucleus. The dashed red lines indicate a region where mitochondria are absent. The white arrowhead indicates sperm annulus. Scale bar = 0.2 μm. (H) TEM images indicate the ultrastructure of the midpiece, principal piece and end piece of WT and Fbxo24 KO sperm flagellum in the cross-sections. Arrows indicate the mitochondria (MI), outer dense fiber (ODF) and axoneme. Scale bar = 0.2 μm. Western blot shows the levels of proteins of mitochondria (I) and axoneme (J) of WT and Fbxo24 KO sperm. GAPDH serves as a loading control.
-
Figure 3—source data 1
Raw western blot for Figure 3I, J.
- https://cdn.elifesciences.org/articles/91666/elife-91666-fig3-data1-v1.zip
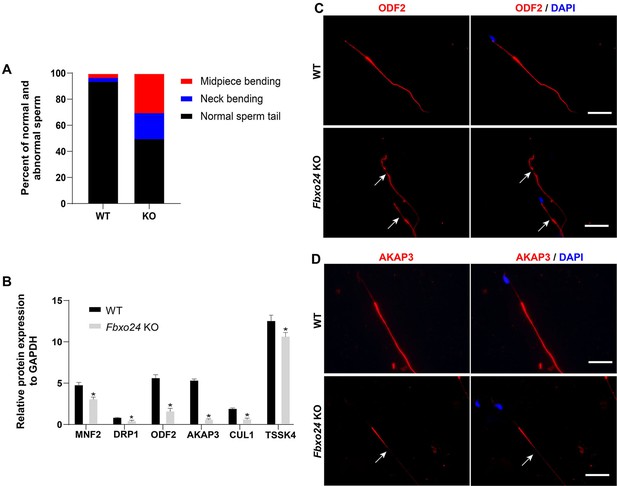
Sperm morphology analysis in FBXO24-deficient mice.
(A) Percentage of morphologically normal and abnormal spermatozoa in wild-type (WT) and Fbxo24 knockout (KO) sperm. (B) Quantification of protein levels of MFN2, DPR1, ODF2, AKAP3, CUL1, and TSSK4 in WT and Fbxo24 KO sperm. Error bars represent mean ± standard deviation (SD), n = 3. *p < 0.05. (C) ODF2 and (D) AKAP3 immunofluorescent signals in the flagellum of WT and Fbxo24 KO sperm. The arrows indicated the week or absent staining of the immunofluorescent signal. Scale bar, 50 µm.
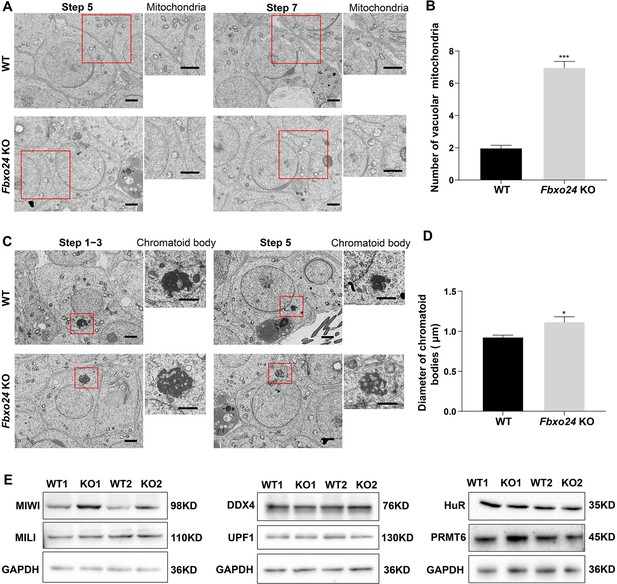
Ablation of FBXO24 affects mitochondria and chromatoid body (CB) architecture in the round spermatids.
(A) Transmission electron microscope (TEM) images show vacuolar mitochondria with disorganized cristae in the round spermatids of Fbxo24 knockout (KO) testes. Right panel insets show higher magnification of mitochondria. Scale bars = 1 μm. (B) Quantification of the number of vacuolar mitochondria. Error bars represent mean ± SD. n = 3. ***p < 0.001. (C) TEM images showing decondensed and enlarged CB with an irregular network in the round spermatids of Fbxo24 KO testes. Right panel insets show a higher magnification of CB. Scale bars = 1 μm. (D) Quantification of size/diameters of CB. Error bars represent mean ± SD. n = 3. *p < 0.05. (E) Western blot analysis expression levels of CB components and PRMT6 in testes from wild-type (WT) and Fbxo24 KO mice at 8 weeks old. GAPDH serves as a loading control.
-
Figure 4—source data 1
Raw western blot for Figure 4E.
- https://cdn.elifesciences.org/articles/91666/elife-91666-fig4-data1-v1.zip
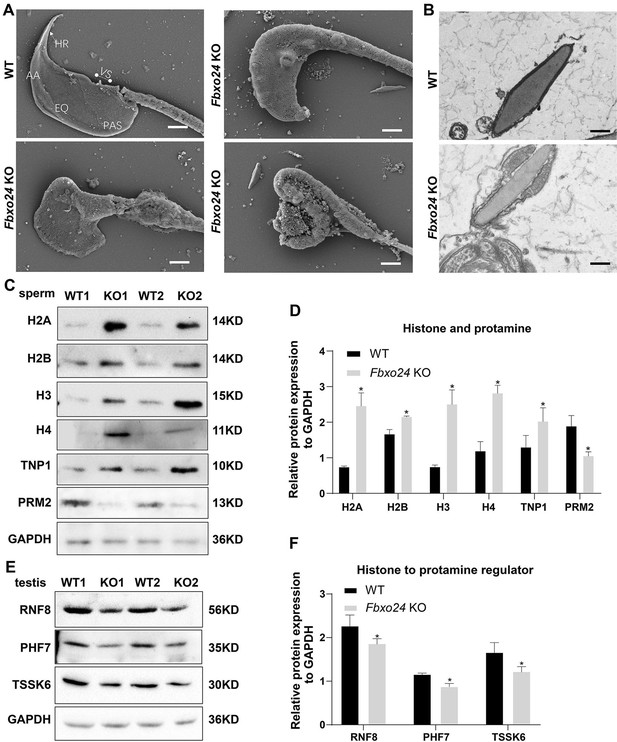
FBXO24 deficiency in mice impairs sperm histone-to-protamine exchange.
(A) Scanning electron microscope (SEM) images show the abnormality of Fbxo24 knockout (KO) sperm head. AA, anterior acrosome; EQ, equatorial segment; PAS, post-acrosomal segment; VS, ventral spur; HR, hook rim. Scale bars = 2 μm. (B) Transmission electron microscopy (TEM) images show the decondensed nucleus (Nu) of Fbxo24 KO sperm. Scale bars = 1 μm. (C) Western blot analysis of the expression of histones (H2A, H2B, H3, and H4), transition proteins (TNP1), and protamines (PRM2) from wild-type (WT) and Fbxo24 KO sperm are shown. GAPDH serves as a loading control. (D) Quantification of protein levels. Error bars represent mean ± standard deviation (SD), n = 3. *p < 0.05. (E) Western blot analysis of the expression of indicated proteins in WT and Fbxo24 KO testis. GAPDH serves as a loading control. (F) Quantification of protein levels. Error bars represent mean ± SD, n = 3. *p < 0.05.
-
Figure 5—source data 1
Raw western blot for Figure 5C, E.
- https://cdn.elifesciences.org/articles/91666/elife-91666-fig5-data1-v1.zip

DNA damage, histone, and transition protein analysis.
(A) DNA damage analysis by TUNEL assay in wild-type (WT) and Fbxo24 knockout (KO) sperm. Scale bar, 50 µm. Percentage of apoptotic cells in WT and Fbxo24 KO sperm. Error bars represent mean ± standard deviation (SD), n = 3. ***p < 0.001. (B) Histone and transition protein analysis in immunoprecipitation. The testis lysate of FBXO24-HA-tagged mice were immunoprecipitated with anti-HA beads. Western blots were used to detect the histone and transition protein expression.
-
Figure 5—figure supplement 1—source data 1
Raw western blot for Figure 5—figure supplement 1B.
- https://cdn.elifesciences.org/articles/91666/elife-91666-fig5-figsupp1-data1-v1.zip

RNA-seq analyses of the round spermatids from FBXO24-deficient testes.
(A) Volcano plot of differentially expressed transcripts in the round spermatids (RS) of Fbxo24 knockout (KO) vs. wild-type (WT) mice. Each red (up-regulation) or blue (down-regulation) dot represents a significantly changed gene. (B) Gene Ontology (GO) term enrichment analysis of down-regulated transcripts of Fbxo24 KO RS. Gene expressions of mitochondrion localization (C) and chromatin organization (D) in RNA-seq analysis. (E) Venn diagrams showing the overlap between down-regulated genes and abnormal alternative splicing genes in Fbxo24 KO RS. (F) Summary of differential splicing evens in Fbxo24 KO RS. The number of each category of alternative splicing is indicated.
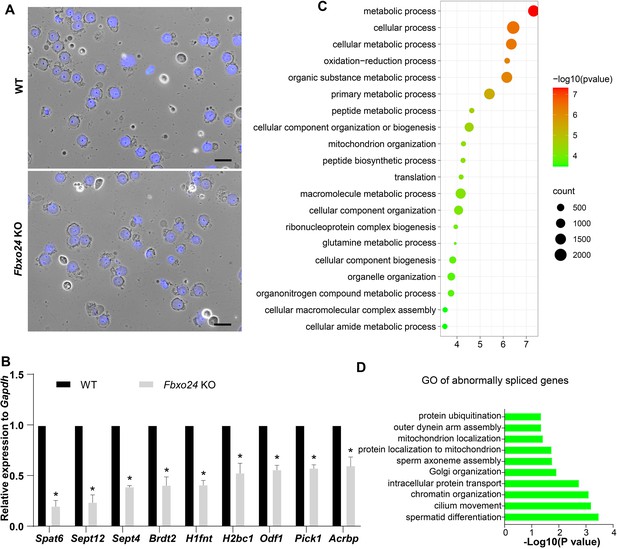
Global gene expression altered in round spermatids of FBXO24-deficient mice.
(A) The purity and morphology of isolated round spermatids can be determined from bright-field images merged with DAPI nuclear staining (blue). Scale bar = 50 μm. (B) qPCR validation of some genes in the testis of wild-type (WT) and Fbxo24 knockout (KO) adult mice. The data are represented as mean ± standard deviation (SD). *p < 0.05. n = 3. (C) Gene Ontology (GO) of the top 20 GO terms of the up-regulated genes in Fbxo24 KO round spermatids. (D) GO of the abnormally spliced genes in Fbxo24 KO round spermatids.

Aberrant alternative splicing of spermiogenesis genes in the round spermatids of FBXO24-deficient mice.
(A) Co-immunoprecipitation analysis of FBXO24 and the splicing regulators (SRSF2, SRSF3, and SRSF9) in Fbxo24-HA-tagged mice testis. Wild-type (WT) testis was used as a negative control. (B) Western blotting analysis of the splicing regulators in WT and Fbxo24 knockout (KO) testis. GAPDH was used as a loading control. (C) Quantification of protein levels. Error bars represent mean ± standard deviation (SD), n = 3. *p < 0.05. Validation of abnormal alternative splicing genes related to (D) mitochondria (Mfn1), (E) flagellum (Zmynd12, Map7d1, and Bbs7), (F) chromatin (Phf7 and Kat5), (G) acrosome (Nucb2), and (H) ubiquitination (Ube2j2). The top panels represent RT-PCR analysis of indicated genes in WT and Fbxo24 RS. Gapdh serves as a loading control. The middle panels show the quantification of percent spliced in (PSI) and alternative sites in RNA-seq. Error bars represent mean ± SD, n = 2. *p < 0.05. The bottom panels represent the schematic diagram of alternative sites exons.
-
Figure 7—source data 1
Raw western blot for Figure 7A, B.
- https://cdn.elifesciences.org/articles/91666/elife-91666-fig7-data1-v1.zip
-
Figure 7—source data 2
Raw RT-PCR gel for Figure 7D–H.
- https://cdn.elifesciences.org/articles/91666/elife-91666-fig7-data2-v1.zip
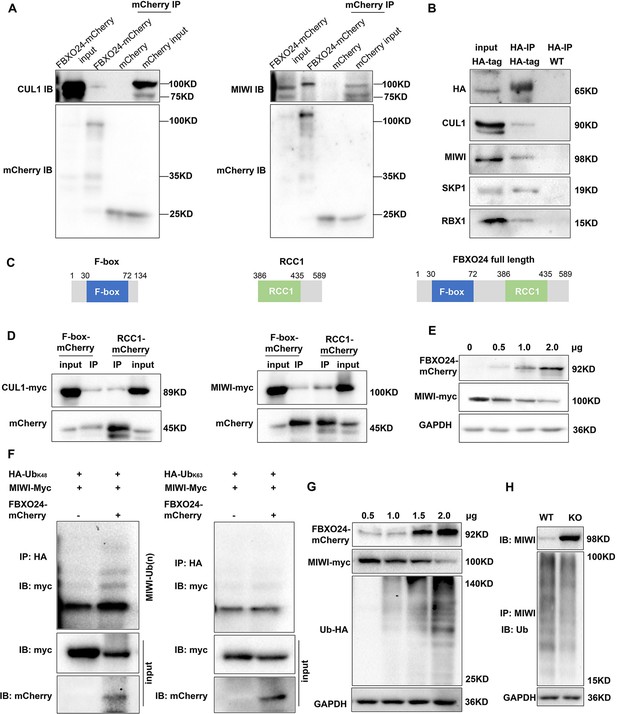
FBXO24 interacts with MIWI and mediates its K48-linked polyubiquitination.
(A) HEK293T cells transfected with empty FBXO24-mCherry or mCherry vector. Anti-mCherry beads were used for immunoprecipitation (IP), and western blots were used to detect the CUL1 (left panel) and MIWI (right panel) expression. (B) The testis lysate of Fbxo24-HA-tagged mice were immunoprecipitated with anti-HA beads. Western blots were used to detect the HA, CUL1, MIWI, SKP1, and RBX1expression. (C) Schematic structures of the truncated FBXO24 protein are shown. Broken boxes show the domain of F-box and regulator of chromosome condensation 1 (RCC1). (D) HEK293T cells were transfected with indicated plasmids. IP was performed using the anti-mCherry antibody. (E) Western blot analysis of HEK293T cells transfected with indicated FBXO24-mCherry and 2 μg MIWI-myc plasmids. The cell lysates were immunoblotted with anti-mCherry and anti-myc antibodies. (F) FBXO24 mediated the ubiquitination of MIWI in the presence of Ub (K48) not Ub (K63). (G) HEK293T cells were transfected with indicated FBXO24-mCherry, 2 μg MIWI-myc, and 2 μg Ub-HA plasmids. The cell lysates were immunoblotted with the anti-mCherry, anti-myc, and anti-HA antibodies. GAPDH serves as a loading control. (H) Ubiquitination analysis of MIWI in the round spermatids of Fbxo24 knockout (KO) mice. The cells were treated with MG132 (10 µM) in the ubiquitination assay.
-
Figure 8—source data 1
Raw western blot for Figure 8A, B, D–H.
- https://cdn.elifesciences.org/articles/91666/elife-91666-fig8-data1-v1.zip
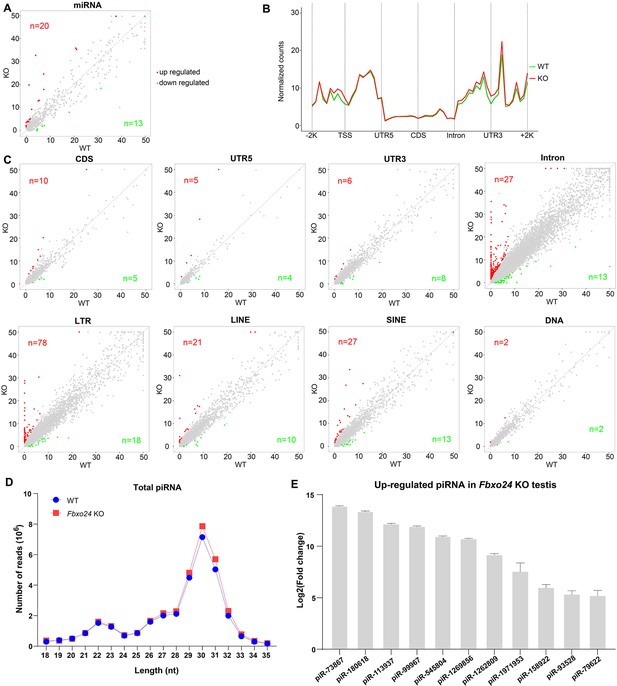
Small RNA-seq analysis of testes from FBXO24-deficient mice.
(A) A scatter plot of differentially expressed miRNA is shown. Red and green dots represent up- and down-regulated miRNA (fold change >2, p < 0.05), respectively. (B) Genomic distribution of piRNA profile in Fbxo24 knockout (KO) vs. wild-type (WT) testis. piRNA levels were examined in each 200-bp interval of a 2-kb region up- and downstream of the annotated genes. (C) Scatter plots of differentially expressed piRNA mapping density (reads/kb) of the coding region (CDS), 5′ and 3′ untranslated region (UTR), and intron, as well as transposable element (TE), including retrotransposon (LTR, LINE, and SINE) and DNA transposon. The piRNA read counts were normalized with miRNA. Red and green dots represent up- and down-regulated piRNA (fold change >2, p < 0.05), respectively. (D) The size distribution of piRNAs in Fbxo24 KO vs. WT testis. (E) The top 10 up-regulated piRNAs in Fbxo24 KO testis exist in MIWI immunoprecipitates of GSM822760 data.
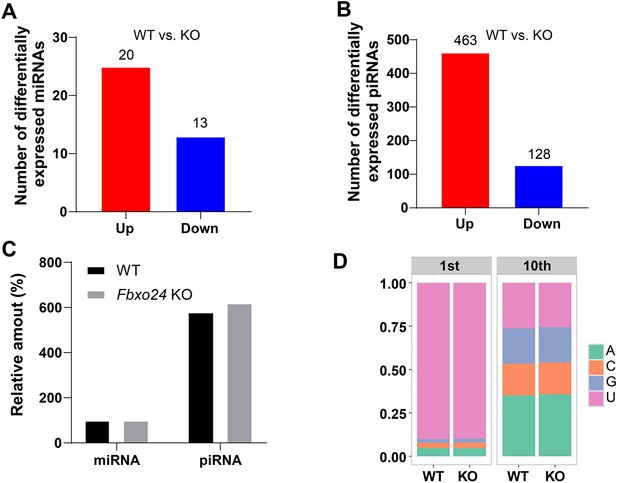
miRNA and piRNA expression analysis in the FBXO24-deficient mice.
(A) Graph bars showing the number of differentially expressed miRNA in the testis of Fbxo24 knockout (KO) mice with 8-week-old (n = 3/group). Significantly regulated genes have a p-value of <0.05 and fold change of >2. The number of up- and down-regulated miRNA is indicated. (B) Graph bars showing the number of differentially expressed piRNA in the testis of Fbxo24 KO mice with 8-week-old (n = 3/group). The number of up- and down-regulated piRNA is indicated. (C) Ratios of total piRNA in KO vs. wild-type (WT) after the normalization with miRNA counts. (D) Ratios of the 1st and 10th nucleotides of the repeat-associated piRNA.

A schematic model shows the FBXO24-mediated post-transcriptional regulation during spermiogenesis.
Fbxo24 interacts with key splicing factors (SRSF2, SRSF3, and SRSF9) to coordinate proper alternative splicing of the target mRNA transcripts involved in spermiogenesis. FBXO24 regulates the architectures of mitochondria and chromatid body through MIWI/piRNA pathway in the round spermatids.
Additional files
-
Supplementary file 1
Identification of FBXO24 interactors by mass spectrometry.
- https://cdn.elifesciences.org/articles/91666/elife-91666-supp1-v1.xlsx
-
Supplementary file 2
Differentially expressed genes in the round spermatids between Fbxo24 KO and wild-type (WT) mice by RNA-seq.
- https://cdn.elifesciences.org/articles/91666/elife-91666-supp2-v1.xlsx
-
Supplementary file 3
Gene Ontology (GO) enrichment of the down-regulated genes in the round spermatids of Fbxo24 KO by RNA-seq.
- https://cdn.elifesciences.org/articles/91666/elife-91666-supp3-v1.xlsx
-
Supplementary file 4
Differentially expressed miRNAs in the testis between Fbxo24 KO and wild-type (WT) mice by small RNA-seq.
- https://cdn.elifesciences.org/articles/91666/elife-91666-supp4-v1.xlsx
-
Supplementary file 5
Differentially expressed piRNAs in the testis between Fbxo24 KO and wild-type (WT) mice by small RNA-seq.
- https://cdn.elifesciences.org/articles/91666/elife-91666-supp5-v1.xlsx
-
Supplementary file 6
The expression of transposable element-derived piRNAs in small RNA-seq data.
- https://cdn.elifesciences.org/articles/91666/elife-91666-supp6-v1.xlsx
-
Supplementary file 7
Primer sequences are used in this study.
- https://cdn.elifesciences.org/articles/91666/elife-91666-supp7-v1.xlsx
-
Supplementary file 8
Antibodies used in this study.
- https://cdn.elifesciences.org/articles/91666/elife-91666-supp8-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/91666/elife-91666-mdarchecklist1-v1.docx





