Early recovery of proteasome activity in cells pulse-treated with proteasome inhibitors is independent of DDI2
Figures
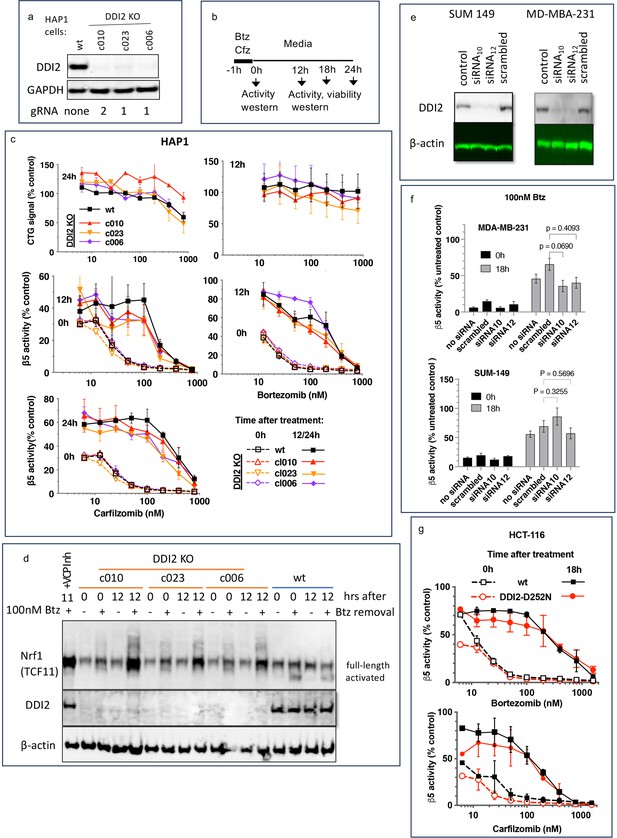
Recovery of proteasome activity is DDI2 independent.
(a) Expression of DDI2 in the CRISPR-generated clones of HAP1 cells used in this work was analyzed by western blot. (b) The experimental setup used in this study. Cells were pulse treated with bortezomib (Btz) or carfilzomib (Cfz) for 1 hr, then cultured in drug-free media for times indicated and analyzed as described. (c) The viability of wt- and DDI2 KO clones of HAP1 cells was measured using CellTiter-Glo, and the inhibition of β5 sites was measured with the Proteasome-Glo assay at times indicated; n=2–5. (d) Knockout of DDI2 inhibits the Nrf1 processing. Western blots of Btz-treated HAP1 cells. The sample in the first lane is wt cells treated with VCP/p97 inhibitor CB-5083 immediately after removal of Btz. VCP inhibitors blocks Nrf1 processing (Radhakrishnan et al., 2014; Sha and Goldberg, 2014; Anderson et al., 2015). (e) MDA-MB-231 and SUM149 cells were analyzed by western blot 72 hr after transfection with DDI2 siRNAs (f) Theβ5 activity in siRNA-transfected SUM149 and MDA-MB-231 was measured using Suc-LLVY-AMC immediately and 18 hr after treatment with 100 nM Btz; n=3. (g) β5 activity was measured in HCT-116 cells with the Proteasome-Glo assay immediately and 18 hr after treatment with PIs; n=2.
-
Figure 1—source data 1
PDF file containing Figure 1a and original full-size western blot membranes (anti-DDI2, anti-GAPDH) with molecular weight markers.
- https://cdn.elifesciences.org/articles/91678/elife-91678-fig1-data1-v1.zip
-
Figure 1—source data 2
Excel file containing data for Figure 1c.
- https://cdn.elifesciences.org/articles/91678/elife-91678-fig1-data2-v1.xlsx
-
Figure 1—source data 3
PDF file containing Figure 1d and original full-size western blot membranes (anti-Nrf1, anti-DDI2, anti-β-actin) with molecular weight markers.
- https://cdn.elifesciences.org/articles/91678/elife-91678-fig1-data3-v1.zip
-
Figure 1—source data 4
PDF file containing Figure 1e and full-size western blot membranes (anti-DDI2, anti-β-actin).
Additional lanes demonstrate that the knockdown of DDI2 is maintained throughout the experiment.
- https://cdn.elifesciences.org/articles/91678/elife-91678-fig1-data4-v1.zip
-
Figure 1—source data 5
Excel file containing data and statistical analysis for Figure 1f.
- https://cdn.elifesciences.org/articles/91678/elife-91678-fig1-data5-v1.xlsx
-
Figure 1—source data 6
Excel file containing data for Figure 1g.
- https://cdn.elifesciences.org/articles/91678/elife-91678-fig1-data6-v1.xls
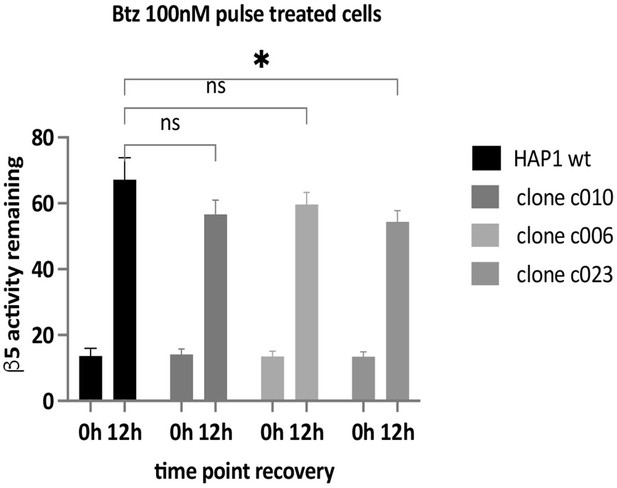
The proteasome activity of the samples used in Figure 1d was measured with Suc-LLVY-AMC; n=9.
-
Figure 1—figure supplement 1—source data 1
Excel file containing data and statistical analysis.
- https://cdn.elifesciences.org/articles/91678/elife-91678-fig1-figsupp1-data1-v1.xlsx
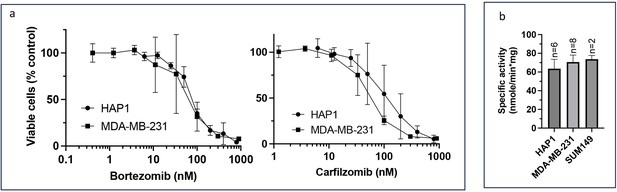
Comparison of proteasome activity and proteasome inhibitor (PI) sensitivity between HAP1, MDA-MB-231, and SUM149 cells.
(a) Cells were treated with PIs for 1 hr, media was shaken off, and cells were cultured in an inhibitor-free fresh media for 48 hr when Alamar Blue assay was performed; n=3–4. See Figure 1 in Weyburne et al., 2017 for a comparison of SUM149 and MDA-MB-231 cells. (b) The β5 proteasome activity was measured using Suc-LLVY-AMC in the cell extracts of untreated cells; n=2-8.
-
Figure 1—figure supplement 2—source data 1
Excel file containing data for both panels.
- https://cdn.elifesciences.org/articles/91678/elife-91678-fig1-figsupp2-data1-v1.xlsx
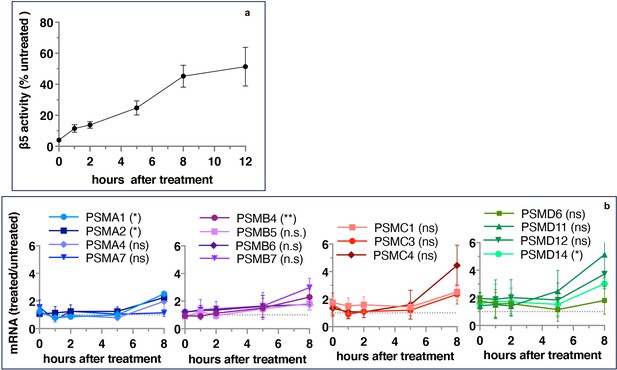
Proteasome activity recovers before upregulation of proteasome gene expression.
Wt-HAP1 cells were pulse-treated with bortezomib (Btz) (100 nM), cultured in a drug-free medium, and analyzed at indicated times. (a) β5 activity was measured using Proteasome-Glo and normalized first to CellTiter-Glo viability data and then to proteasome activity in the mock-treated samples; n=2–5. (b) In a parallel experiment, the mRNA was isolated, and the expression of proteasome genes was quantified using quantitative RT-PCR; n=3. Results of the t-test at 8 hr are in parenthesis.
-
Figure 2—source data 1
Prism file containing data and statistical analysis for both panels.
- https://cdn.elifesciences.org/articles/91678/elife-91678-fig2-data1-v1.xlsx
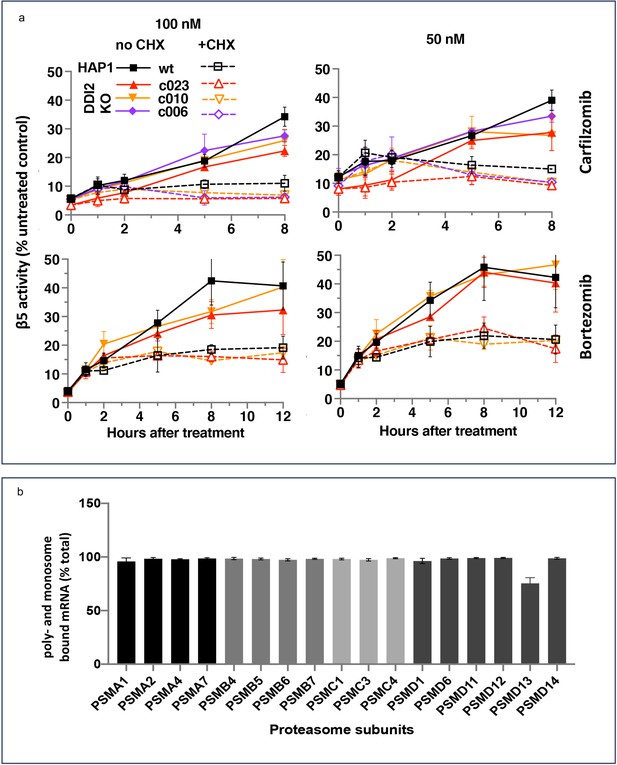
The recovery of proteasome activity requires protein synthesis.
(a) Wt-HAP1 and DDI2 KO cells were treated for 1 hr at indicated concentrations of bortezomib (Btz) and carfilzomib (Cfz) and then cultured in a drug-free media in the absence (solid lines) or presence (dashed lines) of cycloheximide (CHX). The β5 activity was measured using Proteasome-Glo and normalized first to cell viability, which was determined in a parallel experiment using CellTiter-Glo, and then to untreated controls; n=3–4. (b) All proteasome mRNAs are actively translated. mRNA isolated from untreated wt-HAP1 cells were analyzed by polysome profiling. The combined mRNAs in the 80 S and polysomal fractions as a % of the total is shown; n=2.
-
Figure 3—source data 1
Prism file containing data and statistical analysis for Figure 3a.
- https://cdn.elifesciences.org/articles/91678/elife-91678-fig3-data1-v1.xlsx
-
Figure 3—source data 2
Excel file containing data for Figure 3b.
- https://cdn.elifesciences.org/articles/91678/elife-91678-fig3-data2-v1.xlsx

Translation of catalytic subunits is not altered after treatment with inhibitors.
Cells were treated with bortezomib (Btz) for 1 hr, and then cultured in drug-free media. harvested at indicated times and analyzed by polysome profiling and qPCR as in Figure 3b; n=2.
-
Figure 3—figure supplement 1—source data 1
Excel file containing data.
- https://cdn.elifesciences.org/articles/91678/elife-91678-fig3-figsupp1-data1-v1.xlsx
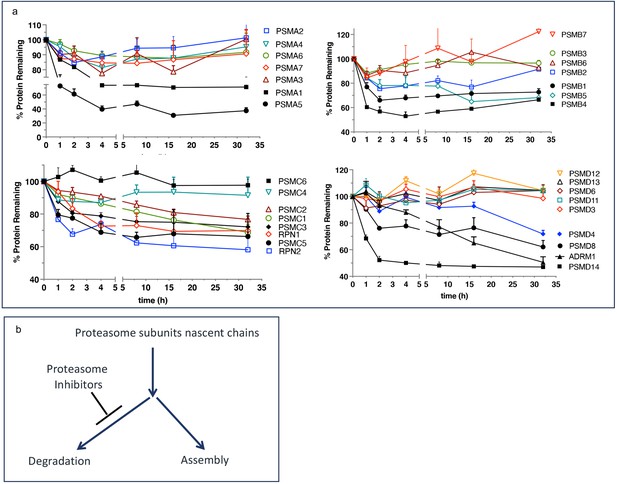
Escape from rapid degradation of nascent subunits can explain rapid recovery of proteasome activity.
(a) Turnover of proteasome subunit in human RPE-1 cells was measured by quantitative mass-spectrometry following 1 hr labeling with heavy isotopes. Data taken from Table S4 in McShane et al., 2016; n=2-3. (b) Proposed model of how nascent proteasome subunits are partitioned between assembly and degradation.
-
Figure 4—source data 1
Prism file containing data from Table S4 in McShane et al., 2016 that was used to create figure.
- https://cdn.elifesciences.org/articles/91678/elife-91678-fig4-data1-v1.txt
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Homo sapiens) | HAP1-wt | Horizon Discovery | RRID:CVCL_Y019, Cat # C631 | Parenteral cell line (clone 631) for DDI2 KO cells below. https://horizondiscovery.com/en/engineered-cell-lines/products/hap1-parental-cell-lines |
| Cell line (Homo sapiens) | HAP1-DDI2 KO, clone 010 | Horizon Discovery | Cat # HZGHC000396c010 | Generated by CRISPR using gRNA:AATAGCTATGGAAGAGGCTC; 41 bp deletion; https://horizondiscovery.com/en/search?searchterm=HZGHC000396c010, |
| Cell line (Homo sapiens) | HAP1-DDI2 KO, clone 023 | Horizon Discovery | Calatogue # HZGHC000182c023 | Generated by CRISPR using gRNA:GCTCGAAGTCGGCGTCGACC; 1 bp insertion; https://horizondiscovery.com/en/search?searchterm=HZGHC000182c023 |
| Cell line (Homo sapiens) | HAP1-DDI2 KO, clone 006 | Horizon Discovery | Cat # HZGHC000182c006 | Genertaed by CRISPR using gRNA GCTCGAAGTCGGCGTCGACC; 4 bp deletion; https://horizondiscovery.com/en/search?searchterm=HZGHC000182c006 |
| Cell line (Homo sapiens) | MDA-MB-231 | ATCC | Cat# HTB-26 | https://www.atcc.org/products/htb-26#detailed-product-information |
| Cell line (Homo sapiens) | SUM149 | BioIVT | RRID:CVCL_3422 | |
| Cell line (Homo sapiens) | HCT-11, wt | https://doi.org/10.7554/eLife.18357 | RRID:CVCL_0291 | A matching wt clone to a mutant below, provided by Murata laboratory |
| cell line (Homo sapiens) | HCT-116, DDI2--D252N | https://doi.org/10.7554/eLife.18357 | Contains CRISPR-generated D252N mutation in the active site of DDI2, provided by Murata laboratory | |
| Transfected construct (Homo sapiens) | DDI2 siRNA10 | Horizon Discovery - Dharmacon | J-032713-10-0050 | Sequences: GGACAUGCUUAAACGGCAC |
| Transfected construct (Homo sapiens) | DDI2 siRNA12 | Horizon Discovery - Dharmacon | J-032713-12-0050 | Sequence: CAAGAAAGGAUUCGUCUGU |
| Transfected construct (Homo sapiens) | Non-targeting pool siRNA | Horizon Discovery - Dharmacon | D-001810-10-20 | Sequences: UGGUUUACAUGUCGACUAA, UGGUUUACAUGUUGUGUGA, UGGUUUACAUGUUUUCUGA, UGGUUUACAUGUUUUCCUA |
| Antibody | Anti-TCF11/NRF1 D5B10 (rabbit mAb) | Cell Signaling | Cat# 8052 S | WB (1:500) |
| Antibody | Anti-GAPDH D4C6R (mouse mAb) | Cell Signaling | Cat# 97166 | WB (1:1000) |
| Antibody | Anti-β-actin 8H10D10 (mouse mAb) | Cell Signaling | Cat #3700 | WB (1:1000) |
| Antibody | Anti-DDI2 (rabbit pAb) | Bethyl Laboratories | Cat# A304-629A | WB (1:5000) |
| Antibody | Anti-rabbit IgG, HRP-linked (goat) | Cell Signaling | Cat#7074 | WB (1:1000) |
| Antibody | Anti-mouse IgG, HRP-linked (goat) | Cell Signaling | Cat#7076 P2 | WB (1:1000) |
| Antibody | Goat anti-Rabbit IgG, Alexa Fluor Plus 647 | Thermofisher - Invitrogen | Cat#A32733 | WB (1:3500) |
| Antibody | Goat anti-Rabbit IgG, Alexa Fluor 680 | Thermofisher - Invitrogen | Cat#A-21076 | WB (1:3500) |
| Antibody | IRDye 800CW Goat anti-Mouse IgG | LI-COR | Cat#926–32210 | WB (1:3500) |
| Commercial assay or kit | DharmaFECT 1 | Horizon Discovery - Dharmacon | T-2001–03 | Transfection reagent for MDA- MB-231 and SUM-149 cells |
| Commercial assay or kit | Proteasome-Glo Assay | Promega | G8622 | Assay for Chymotrypsin-like |
| Commercial assay or kit | CellTiter-Glo Assay | Promega | G7572 | Assay for Cell Viability |
| Commercial assay or kit | Pierce Coomassie Plus (Bradford) Assay | ThermoFisher - Life Technologies | 23238 | Assay for Protein Quantification |
| Commercial assay or kit | TRIzol Reagent | ThermoFisher - Life Technologies | 15596018 | RNA Isolation |
| Commercial assay or kit | High-Capacity cDNA Reverse Transcription kit | Thermofisher - Applied Biosystems | 4368814 | |
| Commercial assay or kit | 2 x SYBR Green Bimake qPCR Master Mix | Selleckchem - Bimake | B21203 | |
| Commercial assay or kit | RNasin Plus Ribonuclease Inhibitor | Promega | N2615 | |
| Chemical compound, drug | Bortezomib | LC Laboratories | AS# 179324-69-7, Cat# B-1408 | Proteasome Inhibitor, |
| Chemical compound, drug | Carfilzomib | LC Laboratories | CAS# 868540-17-4, Cat# C-3022 | Proteasome Inhibitors, |
| Chemical compound, drug | CB-5083 | Cayman Chemicals | CAS# 1542705-92-9, Cat# 19311 | p97 inhibitor, |
| Chemical compound, drug | CHAPS (3-((3-cholamidopropyl) dimethylammonio)–1-propanesulfonate) | Thermo Scientific | CAS# 331717-45-4, Cat # 28300 | Detergent |
| Chemical compound, drug | Cycloheximide | Sigma-Aldrich | CAS# 66-81-9, Cat #C1988 | Protein Synthesis Inhibitor, |
| Chemical compound, drug | Digitonin | GoldBio | CAS# 11024-24-1, Cat# D-180–250 | Detergent |
| Chemical compound, drug | PhosSTOP | Roche | Cat# 4906837001 | Mixture of Phosphatase Inhibitors |
| Chemical compound, drug | Suc-LLVY-AMC | Bachem | CAS# 94367-21-2, Cat # 4011369 | Proteasome substrate |
| Chemical compound, drug | Resazurin sodium salt | Sigma-Aldrich | CAS# 62758-13-8, Cat#R7017 | Alamar Blue Viability Assay |
| Software, algorithm | PRISM | GraphPad | version 10 |
Additional files
-
MDAR checklist
- https://cdn.elifesciences.org/articles/91678/elife-91678-mdarchecklist1-v1.pdf
-
Supplementary file 1
PCR primers used in this work.
- https://cdn.elifesciences.org/articles/91678/elife-91678-supp1-v1.docx





