Photosynthesis: The internal plumbing of algal chloroplasts
A soil-dwelling single-celled green alga called Chlamydomonas might not seem the most promising species to study when exploring the details of photosynthesis. There are more prominent groups of phytoplankton that can photosynthesize, but Chlamydomonas has become a ‘model’ eukaryotic microbe because it is simple to transform genetically and the genome has recently been sequenced.
In algal and plant cells, photosynthesis occurs within chloroplasts. Inside the double membrane that surrounds the chloroplast—known as the chloroplast envelope—light is harvested and converted into biochemical energy by pigment–protein complexes and associated proteins found in membrane structures called thylakoids. Loops of thylakoids merge to form an internal, acidified compartment known as the thylakoid lumen. The thylakoids are bathed in a fluid called the stroma, which contains enzymes that are involved in the fixation of carbon dioxide. Each Chlamydomonas cell has a single, cup-shaped chloroplast.
An enzyme called Rubisco catalyzes the crucial first step in carbon fixation and is found in all photosynthetic organisms. The rate of photosynthesis is limited by the amount of CO2 available to Rubisco: this is a particular problem in water because CO2 diffuses more slowly in water than in air and it is mostly found in the form of bicarbonate (HCO3−). To combat this problem, most microalgae—including Chlamydomonas—have evolved processes called carbon concentrating mechanisms that increase the amount of CO2 around Rubisco (Raven et al., 2014). These mechanisms enhance the efficiency of photosynthesis in three ways: the active transport of bicarbonate into the chloroplast; the conversion of bicarbonate back to CO2 by an enzyme called carbonic anhydrase; and the packaging of Rubisco into a small area to minimize the leakage of CO2.
How does a carbon concentrating mechanism manage to accumulate CO2 concentrations some 50–1000 times higher than are found in the environment? In the past this was a difficult question to answer because details of chloroplast structure were missing. Now, in eLife, Benjamin Engel, Miroslava Schaffer, Wolfgang Baumeister and co-workers at the Max Planck Institute of Biochemistry have combined cryo-focussed ion beam milling and cryo-electron tomography to provide a high-resolution 3D reconstruction of the internal architecture of the Chlamydomonas chloroplast in a near-native state (Engel et al., 2015).
The observations show in exquisite detail how, at the base of the chloroplast, the tips of thylakoid membranes fuse with the inner membrane of the chloroplast envelope. This may allow proteins from outside the chloroplast to enter the thylakoids more easily. Also, openings through the stacks of thylakoids allow the stroma to be continuous throughout the chloroplast. Other insights include details of a structure called the eyespot, which enables the alga to move in response to light.
We already knew that most of the Rubisco was confined to a small area within the stroma—called the pyrenoid—in Chlamydomonas. The pyrenoid is usually surrounded by a dense sheath of non-overlapping starch plates. Extensions of thylakoid membranes, called tubules, pass through the pyrenoid (Figure 1A). This is quite different to the structures in cyanobacteria where Rubisco is found: these structures, which are known as carboxysomes, are separated from the membranes that harvest light, and they have a protein coat and internal scaffold to support the Rubisco and the carbonic anhydrase.
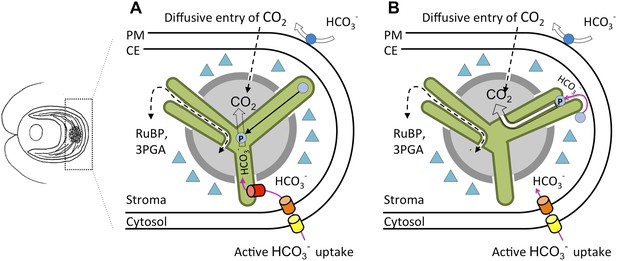
An updated carbon concentrating mechanism for Chlamydomonas.
(A) Existing models of CO2 uptake in Chlamydomonas (Wang et al., 2011) allow for the diffusive entry of CO2, a process facilitated by a carbonic anhydrase, CAH1 (dark blue circle), in the periplasm. There is also a carbon concentrating mechanism that involves the transport of bicarbonate (HCO3−) across the plasma membrane (PM), possibly via the inorganic carbon pumps LCI1 and HLA3 (yellow barrel), and across the chloroplast envelope (CE), possibly via the inorganic carbon pump LCIA (orange barrel). The conversion of bicarbonate to CO2 occurs via CAH3 (light blue circles), a carbonic anhydrase thought to be located inside the lumen of thylakoid tubules (green). When CAH3 is phosphorylated it could relocate within the pyrenoid (light grey). The pump that transfers bicarbonate from the stroma to the lumen (red barrel) has not yet been found. CO2 is prevented from leaking out of the pyrenoid by the starch sheath (dark grey) and LCIB/C protein complexes (triangles). (B) Observations made by Engel et al. demonstrate that minitubules (white) provide a direct route between the stroma and the pyrenoid. This resolves how RuBP, which is the substrate for Rubisco (see text), is channelled to the enzyme, and how 3PGA, which is a product of the reaction between RuBP and CO2, is channelled away. It also raises the intriguing possibility that CAH3 could be external to the thylakoid lumen. When phosphorylated, CAH3 could relocate close to or within the minitubules, also allowing CO2 to be delivered via the minitubules to Rubisco without the need for an additional bicarbonate transporter.
The three-dimensional reconstructions of the pyrenoid show how the tubules extend from thylakoids at the interface of two starch plates, and how they fuse and then merge at the centre of the pyrenoid. Some of these features were visible in early electron micrograph images (Sager and Palade, 1957), but Engel et al. reveal how additional features called minitubules are organised within the tubules that cross the pyrenoid (Figure 1B). The minitubules contain stroma and the latest work clearly shows that they could provide routes for molecules to move from the stroma directly into the centre of the pyrenoid. This would allow substrates and products to move between Rubisco and the other enzymes involved in photosynthesis (Süss et al., 1995), and could reduce the need to transport bicarbonate across the thylakoid membranes, as is required in existing models (Wang et al., 2011).
Engel et al. also reveal how Rubisco is packaged in the pyrenoid. If their proposed model of close-packing in hexagonal shapes is exact, the spacing of Rubisco complexes leaves room for molecules of between 5 kDa and ~50 kDa, depending on the packaging scenario. The latter is sufficient space for a Rubisco linker protein like the one needed inside the carboxysomes of bacteria, but it is not clear whether such proteins also exist in algae (Long et al., 2007).
We can now superimpose these fine details onto existing models of how the carbon concentrating mechanism in Chlamydomonas is organized (Figure 1A). When the mechanism is switched on, bicarbonate could be converted by a stromal carbonic anhydrase within or adjacent to the minitubules (Figure 1B) to allow CO2 to be released at the heart of the pyrenoid.
This new model poses a number of questions. How does Rubisco move to the pyrenoid when the carbon concentrating mechanism is switched on? What is the trigger that modifies the thylakoid membranes to make the tubules as well as multiple minitubules? Why do the starch plates form and surround the pyrenoid? Efforts are being made to engineer elements of the carbon concentrating mechanism from cyanobacteria into higher plants (Lin et al., 2014; McGrath and Long, 2014), but what modifications would be needed to incorporate a mechanism from algae instead?
Undoubtedly, the many genetic techniques that can be used to study Chlamydomonas (Zhang et al., 2014) will soon resolve these questions in the case of this particular alga. However, in the best traditions of product placement, other forms of pyrenoid are available in different species, including phytoplankton and one lineage of primitive land plants. Understanding the structure of chloroplasts and the regulation of carbon concentrating mechanisms in these organisms will continue to provide additional challenges for the future.
References
-
Analysis of carboxysomes from the Synechococcus PCC7942 reveals multiple Rubisco complexes within carboxysomal proteins CcmM and CcaAJournal of Biological Chemistry 282:29323–29335.https://doi.org/10.1074/jbc.M703896200
-
Energy costs of carbon dioxide concentrating mechanisms in aquatic organismsPhotosynthesis Research 121:111–124.https://doi.org/10.1007/s11120-013-9962-7
-
Structure and development of the chloroplast in Chlamydomonas I. The normal green cellJournal Biophysical and Biochemical Cytology 3:463–488.https://doi.org/10.1083/jcb.3.3.463
-
In Situ Association of Calvin Cycle Enzymes, Ribulose-1,5-Bisposphate Carboxyalsae/Oxygenase Activase, Ferredoxin-NADP+ Reductase, and Nitrite Reductase with Thylakoid and Pyrenoid Membranes of Chlamydomonas reinhardtii Chloroplasts as Revealed by Immunoelectron MicroscopyPlant Physiology 107:1387–1397.https://doi.org/10.1104/pp.107.4.1387
Article and author information
Author details
Publication history
- Version of Record published: January 13, 2015 (version 1)
Copyright
© 2015, Meyer and Griffiths
This article is distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use and redistribution provided that the original author and source are credited.
Metrics
-
- 3,580
- views
-
- 227
- downloads
-
- 5
- citations
Views, downloads and citations are aggregated across all versions of this paper published by eLife.
Download links
Downloads (link to download the article as PDF)
Open citations (links to open the citations from this article in various online reference manager services)
Cite this article (links to download the citations from this article in formats compatible with various reference manager tools)
Further reading
-
- Structural Biology and Molecular Biophysics
Roco proteins entered the limelight after mutations in human LRRK2 were identified as a major cause of familial Parkinson’s disease. LRRK2 is a large and complex protein combining a GTPase and protein kinase activity, and disease mutations increase the kinase activity, while presumably decreasing the GTPase activity. Although a cross-communication between both catalytic activities has been suggested, the underlying mechanisms and the regulatory role of the GTPase domain remain unknown. Several structures of LRRK2 have been reported, but structures of Roco proteins in their activated GTP-bound state are lacking. Here, we use single-particle cryo-electron microscopy to solve the structure of a bacterial Roco protein (CtRoco) in its GTP-bound state, aided by two conformation-specific nanobodies: NbRoco1 and NbRoco2. This structure presents CtRoco in an active monomeric state, featuring a very large GTP-induced conformational change using the LRR-Roc linker as a hinge. Furthermore, this structure shows how NbRoco1 and NbRoco2 collaborate to activate CtRoco in an allosteric way. Altogether, our data provide important new insights into the activation mechanism of Roco proteins, with relevance to LRRK2 regulation, and suggest new routes for the allosteric modulation of their GTPase activity.
-
- Developmental Biology
- Structural Biology and Molecular Biophysics
A crucial event in sexual reproduction is when haploid sperm and egg fuse to form a new diploid organism at fertilization. In mammals, direct interaction between egg JUNO and sperm IZUMO1 mediates gamete membrane adhesion, yet their role in fusion remains enigmatic. We used AlphaFold to predict the structure of other extracellular proteins essential for fertilization to determine if they could form a complex that may mediate fusion. We first identified TMEM81, whose gene is expressed by mouse and human spermatids, as a protein having structural homologies with both IZUMO1 and another sperm molecule essential for gamete fusion, SPACA6. Using a set of proteins known to be important for fertilization and TMEM81, we then systematically searched for predicted binary interactions using an unguided approach and identified a pentameric complex involving sperm IZUMO1, SPACA6, TMEM81 and egg JUNO, CD9. This complex is structurally consistent with both the expected topology on opposing gamete membranes and the location of predicted N-glycans not modeled by AlphaFold-Multimer, suggesting that its components could organize into a synapse-like assembly at the point of fusion. Finally, the structural modeling approach described here could be more generally useful to gain insights into transient protein complexes difficult to detect experimentally.





