Ternatin and improved synthetic variants kill cancer cells by targeting the elongation factor-1A ternary complex
Figures
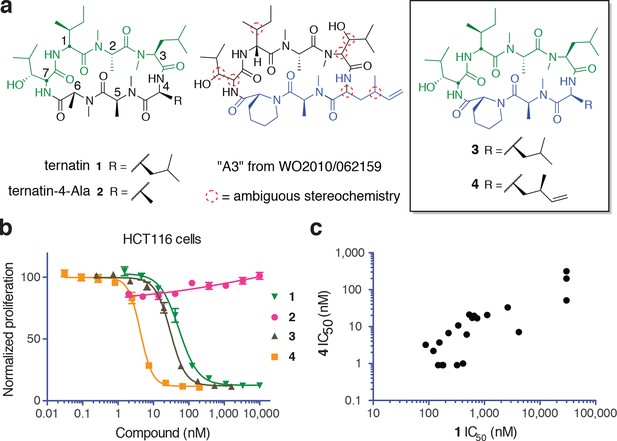
New ternatin variants inspired by Aspergillus-derived cyclic peptides.
(a) Design of ternatin variants 3 and 4, based on the partially elucidated structure of A3. (b) Effect of cyclic peptides 1–4(three-fold dilutions) on HCT116 cell proliferation over 72 hr. (c) Compounds 1 and 4 were tested against a panel of 21 cancer cell lines. Shown is a scatter plot comparing IC50 values of 1 and 4 for each cell line (Spearman correlation, 0.84, p < 0.0001).
-
Figure 1—source data 1
Antiproliferative activity of 1 and 4 against cancer cell lines.
- https://doi.org/10.7554/eLife.10222.004

Epimers 4 and 4b have similar antiproliferative activity.
(a) Chemical structure of 4b, containing (2S,4S)-dehydro-homoleucine. (b) Antiproliferative activity of 4 and epimer 4b toward HCT116 colorectal cancer cells (4b IC50 7.4 ± 1.3 nM).
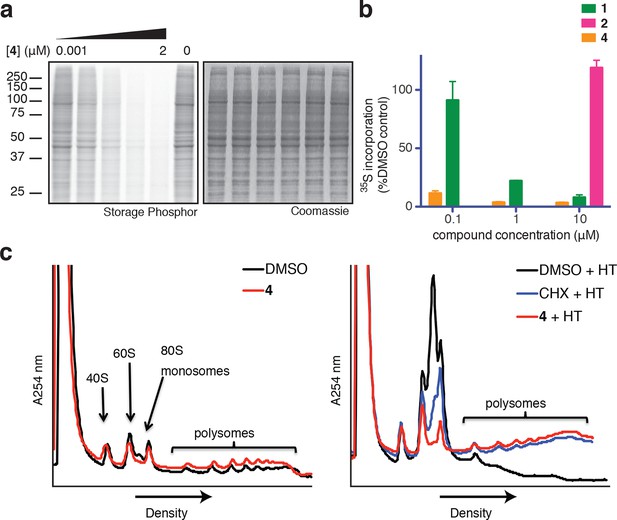
Ternatins inhibit global protein synthesis.
(a) HCT116 cells were treated with compound 4 (five-fold dilutions) for 5 hr before labeling with 35S-Met for 1 hr. Cell lysates were separated by gel electrophoresis. Newly synthesized and total proteins were visualized by autoradiography and Coomassie staining. (b) Cells were treated as in (a), and 35S-labeled proteins were quantified by liquid scintillation counting after TCA precipitation (mean ± SEM, n = 3). (c) Left panel: HeLa cells were treated with 4 (5 μM) or DMSO for 20 min. Right panel: HeLa cells were treated with 4 (5 μM), cycloheximide (CHX, 100 μg/mL), or DMSO for 15 min, followed by harringtonine (HT, 2 μg/mL) for 20 min. After compound treatment, lysates were fractionated on 10–50% sucrose density gradients with absorbance detection at 254 nm.
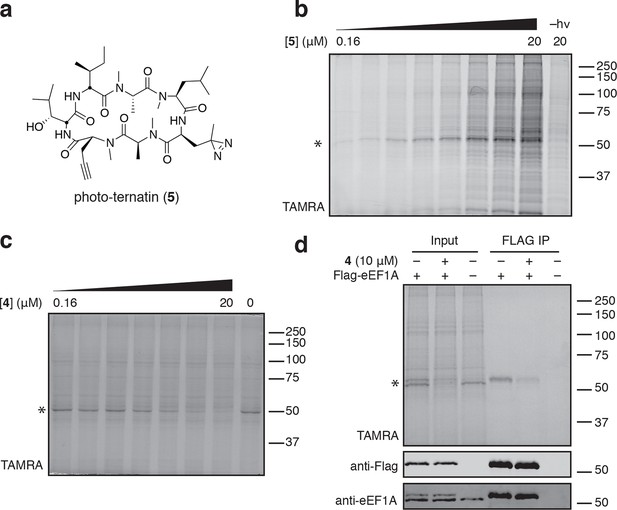
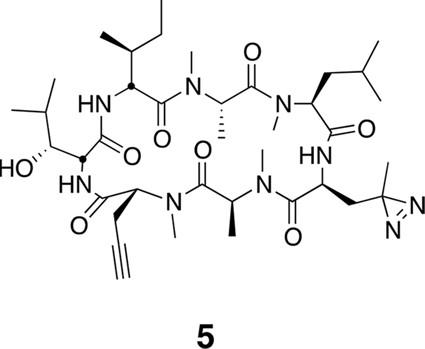
Photo-affinity labeling reveals eEF1A as a direct ternatin target.
(a) Clickable photo-affinity probe 5. (b) HEK293T cell lysates were treated with 5 (two-fold dilutions) for 20 min at room temp followed by UV irradiation (355 nm, 1000 W, 90 s). A control sample with 20 μM 5 was not irradiated. Samples were subjected to click chemistry with TAMRA-azide, separated by gel electrophoresis, and scanned for in-gel fluorescence. (c) Cell lysates were treated with increasing concentrations of 4 for 10 min before adding 5 (2 μM) for 20 min, UV irradiation, and processing as in (b). (d) HEK293T cells were transfected with Flag-eEF1A. Lysates were treated with probe 5 (2 μM) ± 4 and photolyzed as in (b), then immunoprecipitated with magnetic anti-Flag beads. Samples were eluted with SDS, subjected to click chemistry with TAMRA-azide, and analyzed by in-gel fluorescence scanning and Western blotting. Coomassie-stained gels corresponding to (b) and (c) are shown in Figure 3—figure supplement 1.
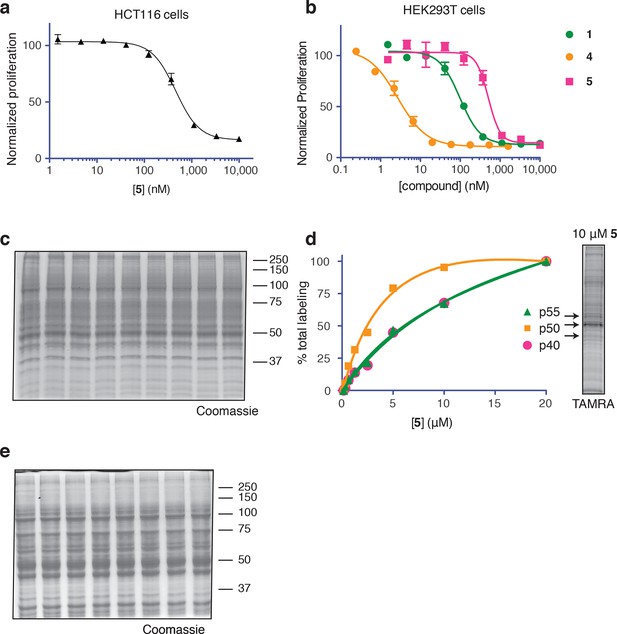
Ternatin photo-affinity probe specifically labels a 50-kDa protein.
(a) 72-hr cell proliferation assay with serial dilutions of 5. (b) 72-hr HEK293T cell proliferation assay. (c) Coomassie-stained gel corresponding to Figure 3b. (d) Dependence of photo-crosslinking intensity on the concentration of photo-ternatin 5 based on in-gel TAMRA fluorescence (Figure 3b). For each of the indicated bands (p40, p50, and p55), the background-corrected fluorescence intensity at a given concentration of 5 was normalized to its intensity in the sample containing 20 μM 5. Labeling of p50, but not p40 or p55, achieves saturation at ~10 μM 5. (e) Coomassie-stained gel corresponding to Figure 3c.
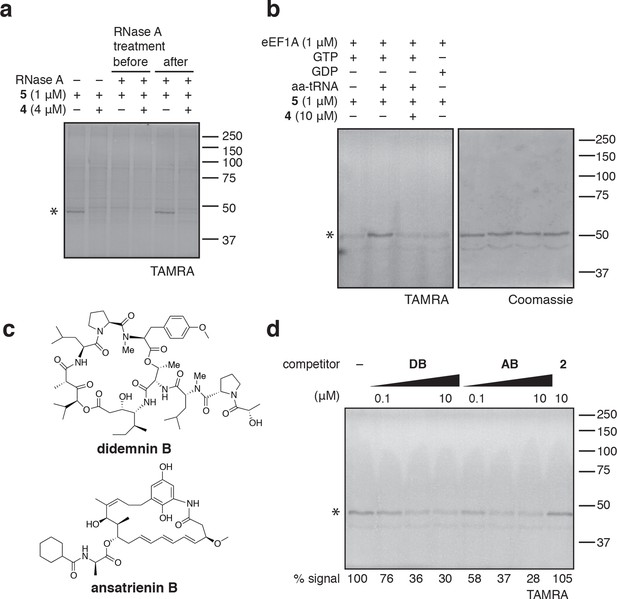
Photo-ternatin 5 binds specifically to the eEF1A ternary complex.
(a) HEK293T cell lysates were treated with RNase A for 20 min before (lanes 3–4) or after (lanes 5–6) incubation with 5 and UV irradiation. (b) Purified eEF1A was incubated with GTP ± Phe-tRNA or GDP for 30 min at room temp. Reactions were treated with DMSO or 4 for 10 min, then 5 for 20 min, photolyzed and processed as in Figure 3b. (c) Translation elongation inhibitors didemnin B (DB) and ansatrienin B (AB). (d) A solution of eEF1A, GTP, and Phe-tRNA was incubated with the indicated compound (DB and AB: 0.1, 1.0, 10 μM) for 10 min before 20-min treatment with 5 (1 μM), followed by UV irradiation and processing as in Figure 3b. Coomassie-stained gels corresponding to (a) and (d) are shown in Figure 4—figure supplement 1.

Coomassie-stained gels corresponding to (a) Figure 4a, and (b) Figure 4d.
In (b), the major 50-kDa band is eEF1A and the minor ~45-kDa band is an unknown contaminant.
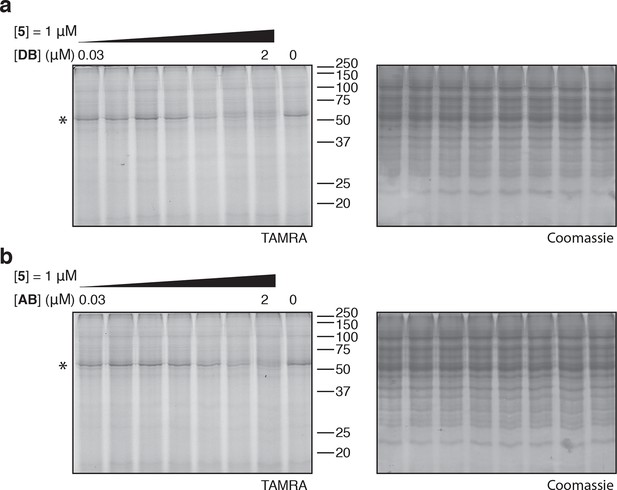
Photo-affinity labeling of HEK293T cell lysates by photo-ternatin 5 in the presence of increasing concentrations of (a) didemnin B (DB), or (b) ansatrienin B (AB).
https://doi.org/10.7554/eLife.10222.011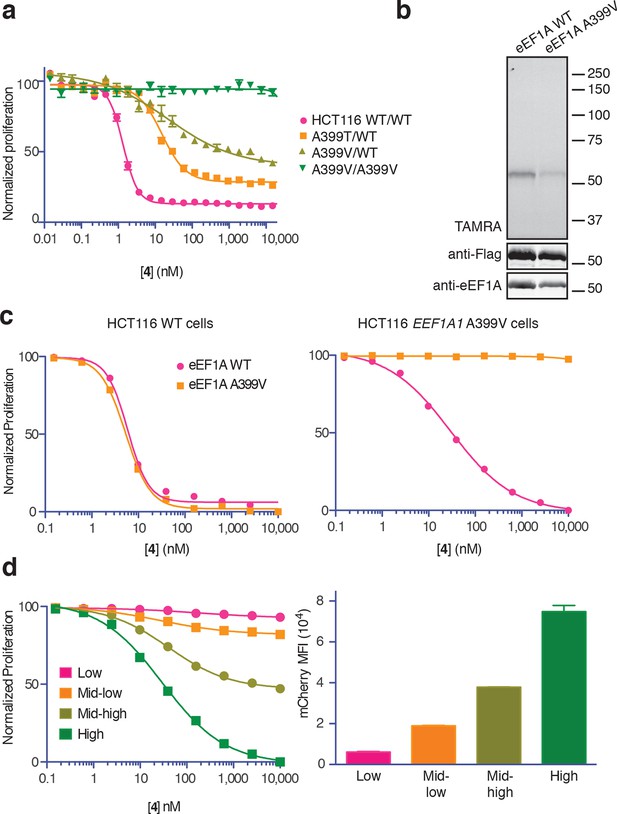
Ala399 mutation in EEF1A1 confers resistance to 4.
(a) Effect of 4 (two-fold dilutions) on proliferation of WT and EEF1A1-mutant HCT116 cells over 72 hr. (b) HEK293T cells were transfected with WT or A399V Flag-eEF1A. Lysates were treated with probe 5 and processed as described in Figure 3d. (c) WT HCT116 cells (left) or cells homozygous for A399V EEF1A1 (right) were transduced with a bicistronic lentiviral vector encoding eEF1A (WT or A399V) and mCherry. Cells were labeled with carboxyfluorescein succinimidyl ester (CFSE), treated with 4 for 72 hr (four-fold dilutions), and analyzed by FACS. Proliferation was assessed by CFSE dilution in high mCherry-expressing cells (mean fluorescence intensity, MFI = 6.7–7.4 × 104). CFSE histograms are shown in Figure 5—figure supplement 1. (d) Homozygous A399V EEF1A1 HCT116 cells were transduced with WT eEF1A/mCherry as described in (c). Antiproliferative effects of 4 (left) were analyzed in cells gated according to the indicated mCherry mean fluorescence intensity (right).
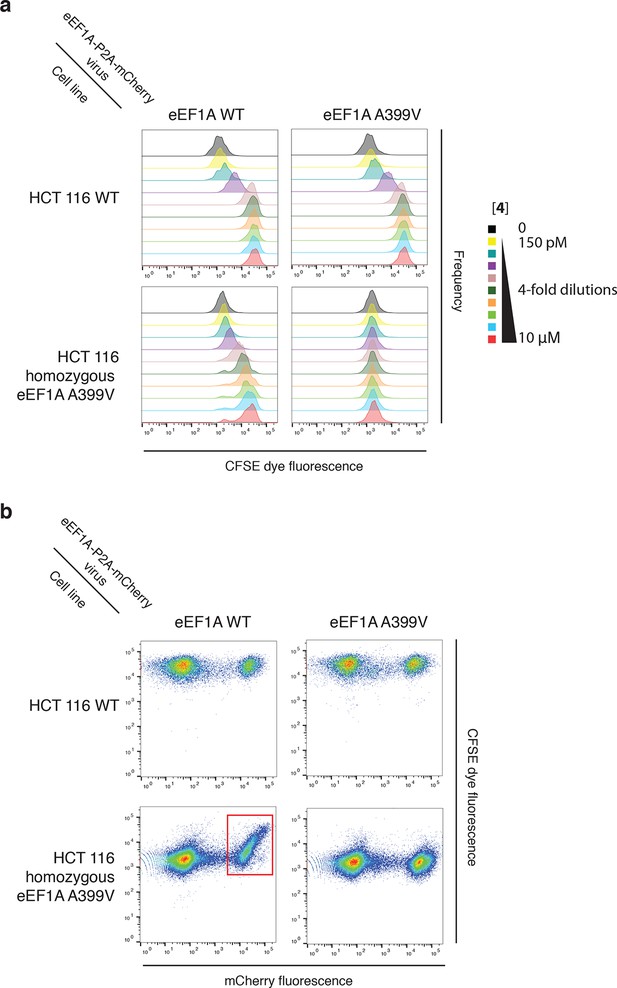
Ternatin sensitivity is genetically dominant.
WT HCT116 cells (top row) or cells homozygous for A399V EEF1A1 (bottom row) were transduced with a bicistronic lentiviral vector encoding WT eEF1A (left column) or A399V eEF1A (right column) linked to mCherry by the ribosome-skipping P2A peptide. Cells were labeled with carboxyfluorescein succinimidyl ester (CFSE), treated with increasing concentrations of 4 for 72 hr, and analyzed by FACS. (a) CFSE histograms for high mCherry cells (mCherry MFI = 6.7–7.4 × 104) were used to generate dose-response curves in Figure 5c. (b) Scatter plots show mCherry and CFSE fluorescence of single cells treated with 10 μM 4. Note the correlation between mCherry and CFSE fluorescence among cells homozygous for A399V EEF1A1 and transduced with WT eEF1A (bottom left plot, red box). In this group, only those cells ectopically expressing high levels of WT eEF1A (indicated by high mCherry fluorescence) fail to proliferate in the presence of 10 μM 4 (indicated by high CFSE fluorescence).
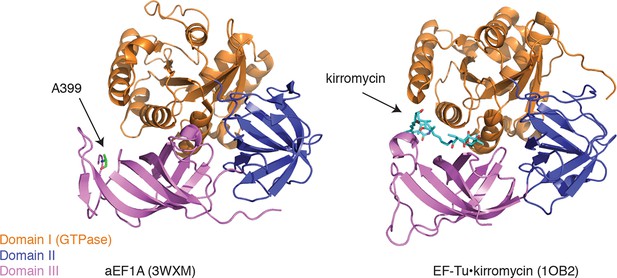
Ternatin may inhibit eEF1A by a mechanism related to kirromycin inhibition of EF-Tu.
Left: crystal structure of archaeal EF1A (PDB code: 3WXM, 51% identity with human eEF1A), showing the location of A399 (human numbering) on the surface of domain III. Right: crystal structure of EF-Tu (PDB code: 1OB2, 25% sequence identity with human eEF1A), showing the kirromycin binding site at the interface of domain I and III.
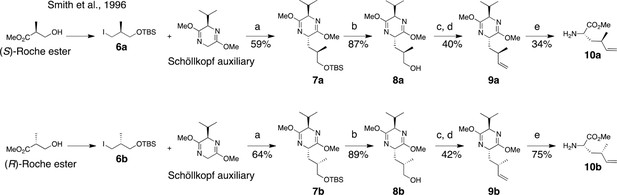
Synthesis of (2S, 4R)- and (2S, 4S)-methyl 2-amino-4-methylhex-5-enoate (dehydro-homoleucine).
Reagents and conditions: (a) nBuLi, THF, –78°C; (b) TBAF, THF, 0°C; (c) Dess-Martin periodinane, CH2Cl2; (d) MePPh3Br, nBuLi, THF, 0°C; (e) TFA, H2O/MeCN.
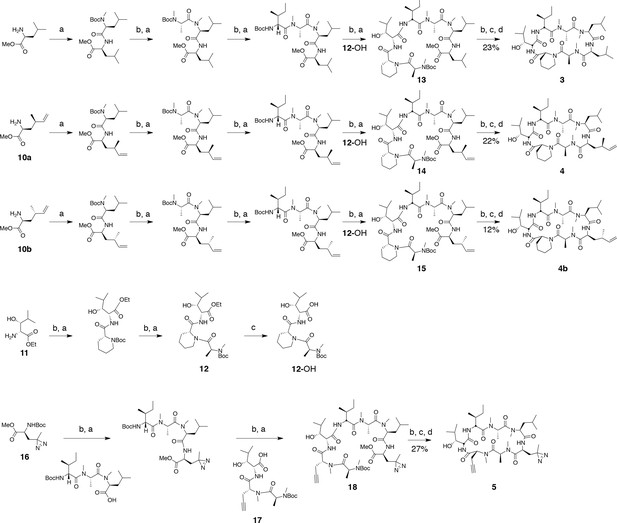
Synthesis of ternatin variants.
Reagents and conditions: (a) HATU, DIPEA, DCM/DMF; (b) 2 M HCl, MeOH, 30°C; (c) 1 N LiOH, H2O/THF; (d) HATU, DIPEA, DMF.
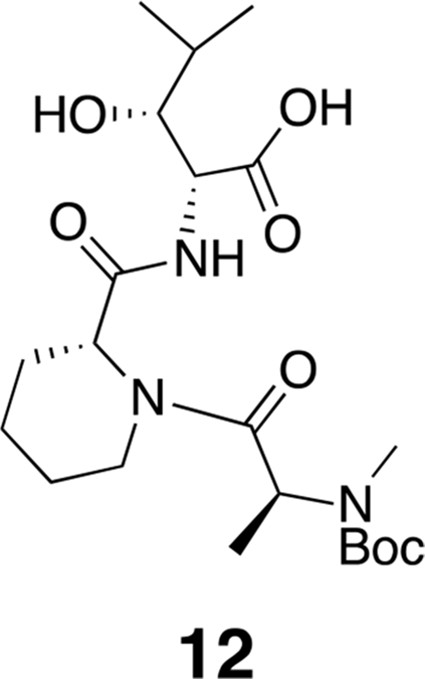
Compound 12.
https://doi.org/10.7554/eLife.10222.017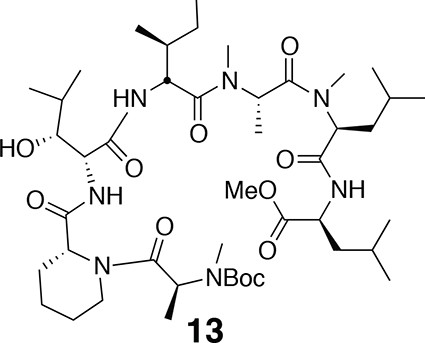
Compound 13.
https://doi.org/10.7554/eLife.10222.018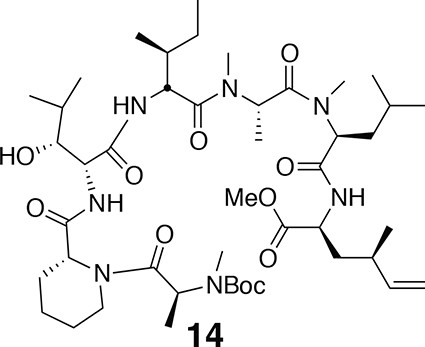
Compound 14.
https://doi.org/10.7554/eLife.10222.019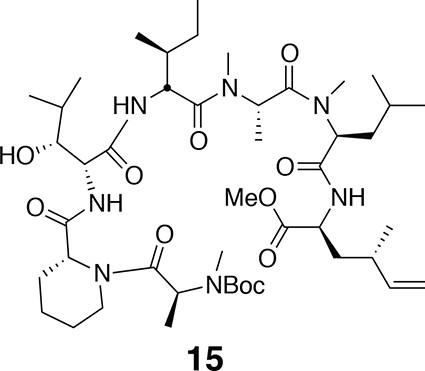
Compound 15.
https://doi.org/10.7554/eLife.10222.020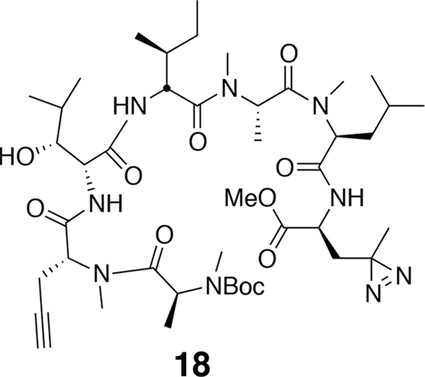
Compound 18.
https://doi.org/10.7554/eLife.10222.021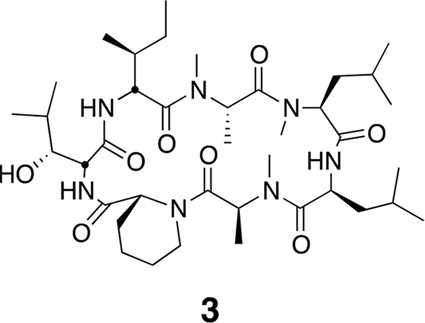
Compound 3.
https://doi.org/10.7554/eLife.10222.022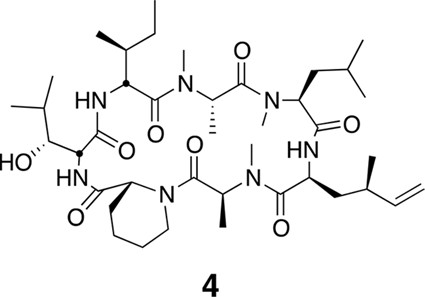
Compound 4.
https://doi.org/10.7554/eLife.10222.023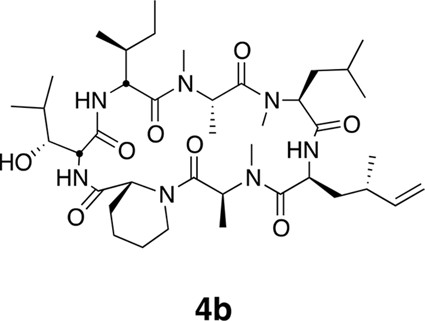
Compound 4b.
https://doi.org/10.7554/eLife.10222.024