The E3 ligase Ubr3 regulates Usher syndrome and MYH9 disorder proteins in the auditory organs of Drosophila and mammals
Figures
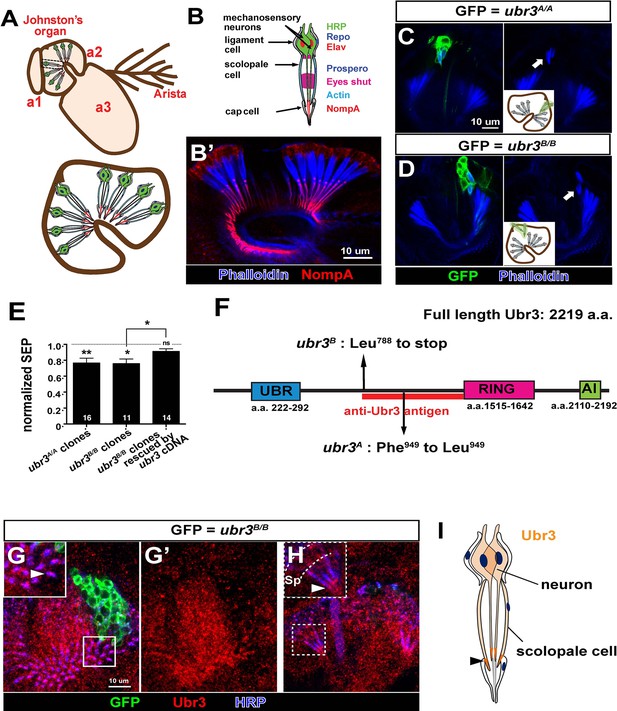
ubr3 regulates auditory organ development in Drosophila.
(A) The structure of the Drosophila auditory organ, Johnston’s organ. The tips of the neuronal cilia are anchored to the cuticle of the third antennal segment by a dendritic cap containing an extracellular glycoprotein, NompA (B) A single scolopidium (corresponding to the box in A) shows the markers used to label various structures and cells in the scolopidium. (B’) Immunolabeling of Johnston’s organ with NompA (red) and phalloidin (actin, blue). (C–D) Pupal Johnston’s organs bearing ubr3 mutant clones were stained with phalloidin to label the actin bundles of scolopale cells. Some ubr3 mutant cells (labeled by GFP) exhibit scolopidia detached from the apical junction of Johnston’s organ (arrows). (E) Extracellular electrophysiological recordings in flies bearing ubr3 mutant clones in Johnston’s organ (left 2 columns) and in flies bearing ubr3 cDNA rescued mutant clones (right column). The data are normalized to flies heterozygous for the corresponding mutations. Numbers of flies recorded are shown in the columns. Error bars show SEM. Statistical symbols show results of t-tests with Welch’s correction as needed (*p<0.05; **p<0.01; ns, not significant). (F) Schematic diagram showing the conserved domains of the Ubr3 protein and the molecular lesions (Phe949 > Leu and Leu788 > STOP) identified in the ubr3A and ubr3B alleles respectively. The red bar shows the epitope used to generate anti-Ubr3 antibody. (G–G’) A single confocal cross-section shows co-immunolabeling of Johnston’s organ with anti-Ubr3 (red) and HRP (neurons, in blue). ubr3B/B mutant clones were generated and labeled with GFP. Ubr3 protein is localized to neuronal cilia marked by an arrowhead. (H) A longitudinal section of Johnston’s organ labeled by anti-Ubr3 (red) and HRP (blue). Ubr3 localizes not only to neuronal cell bodies but faint expression is also seen in cilia. Arrowhead labels enriched Ubr3 proteins in apical ciliary tips. Scolopale cell bodies (Sp) are outlined by dashed lines. (I) Diagram shows distribution of Ubr3 proteins in Johnston’s organ, including its enrichments in the apical tips of the neuronal cilia and the scolopale cells (arrowhead).
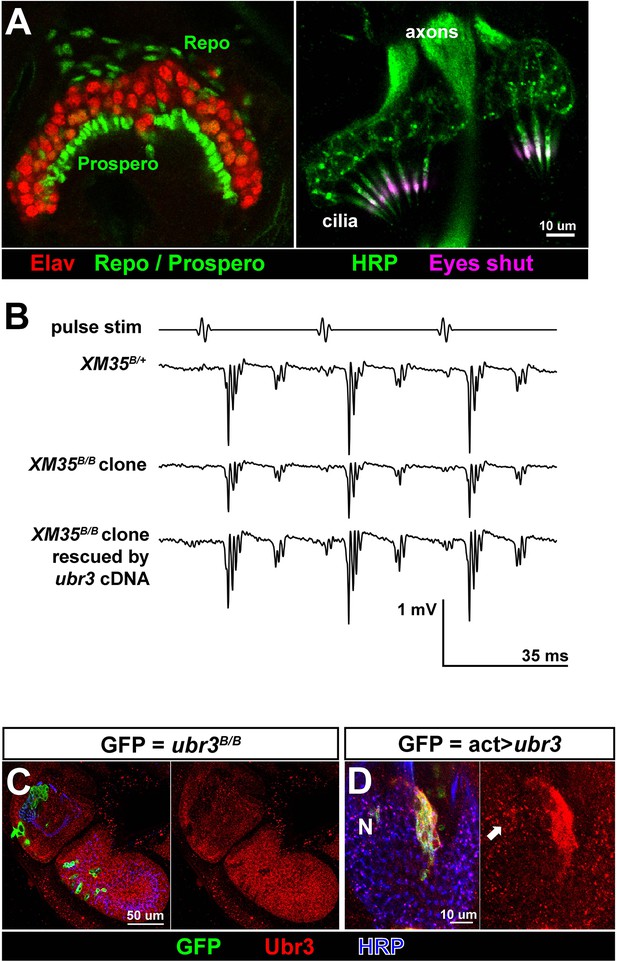
Ubr3 is required for normal development of Johnston’s organ.
(A) Immunolabeling of wild type pupal Johnston’s organs with different markers. (B) Sample traces of sound evoked potential (SEPs) from flies of indicated genotypes. (C) Immunolabeling of the second and third antenna segments of a pupal wild type fly with anti-Ubr3 antibody (red) and HRP (neurons, blue). (D) Immunolabeling of a pupal Johnston’s organ bearing Ubr3 over-expressing clones (labeled by GFP, green) with anti-Ubr3 antibody (red) and anti-HRP (neurons, blue). Ubr3 proteins are present in neuronal cell bodies (N), indicated by arrow.
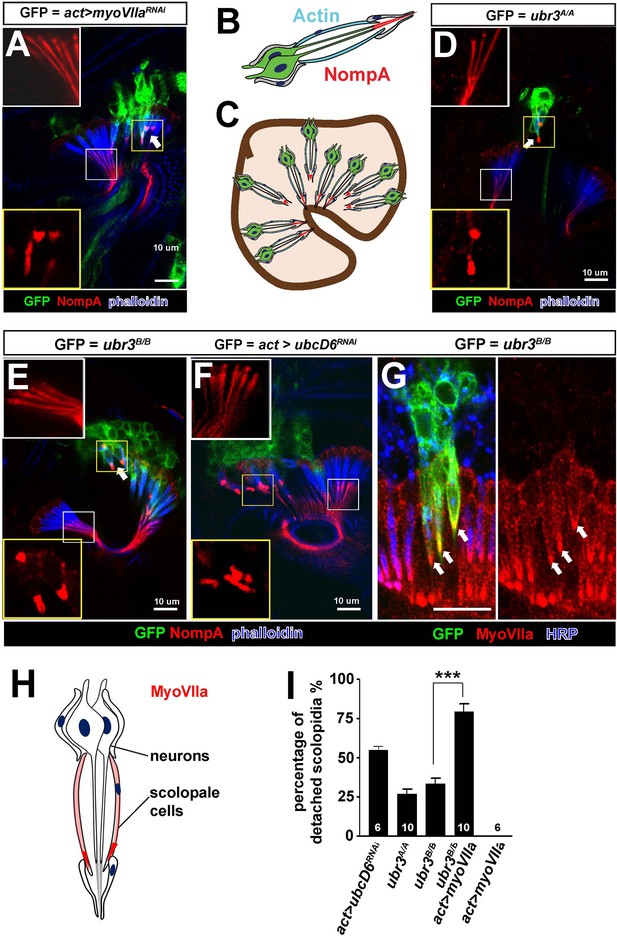
Ubr3 genetically interacts with MyoVIIa.
(A) The normal filamentous structure of NompA in the apical junction of wild-type cells (white box) collapses into puncta in the detached scolopidia in flies in which myoVIIa is knocked down (yellow box). Arrow indicates detached scolopidia. (B) A diagram shows actin (cyan) and NompA (red) in a single scolopidium. (C) A diagram illustrates the detachment of scolopidia and altered NompA pattern. (D–E) The normal filamentous structure of NompA in the apical junction of wild-type cells (white boxes) collapses into puncta in the detached scolopidia in ubr3 mutant cells (labeled by GFP) (yellow boxes). (F) The normal filamentous structure of NompA in the apical junction of wild-type cells (white box) collapses into puncta in the detached scolopidia in cells over-expressing ubcD6 RNAi construct (labeled by GFP) (yellow box). (G) Immunolabeling of Johnston’s organ with ubr3 mutant clones (marked by GFP, green) by anti-HRP (neurons, blue) and anti-MyoVIIa antibody (red). Arrows indicate detached ubr3 mutant scolopidia. (H) A diagram shows localization of MyoVIIa (red) in neuronal cilia and scolopale cells. (I) Quantification of detached scolopidia in the ubr3A/A and ubr3B/B clones, ubcD6 RNAi clones, and wild type or mutant clones over-expressing myoVIIa. Error bars show SEM. Numbers of flies quantified are shown in the columns. (***p<0.001)
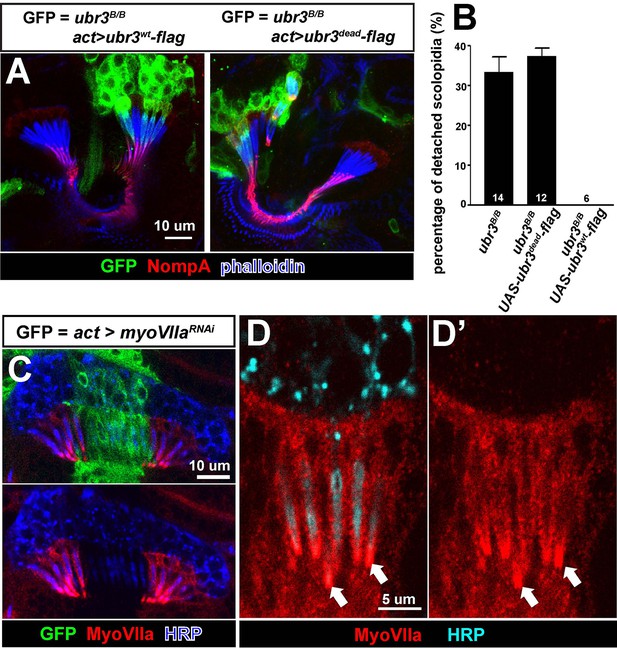
ubr3 mutants phenocopy myoVIIa mutants.
(A) Immunolabeling of pupal Johnston’s organs bearing ubr3 mutant clones over-expressing wild type Ubr3 or E3 ligase-dead form of Ubr3 (labeled by GFP, green) with anti-NompA antibody (red) and phalloidin (actin, blue). (B) Quantification of detached scolopidia in cells with indicated genotypes in Johnston’s organ. Numbers of flies quantified are shown in the columns. (C) Immunolabeling of a pupal Johnston’s organ bearing myoVIIa RNAi expressing clones (labeled by GFP, green) with anti-MyoVIIa antibody (red) and anti-HRP (neurons, blue). (D–D’) Enlargement of the ciliary region of the scolopidia from a wild type Johnston’s organ, labeled by anti-MyoVIIa (red) and anti-HRP (cyan).
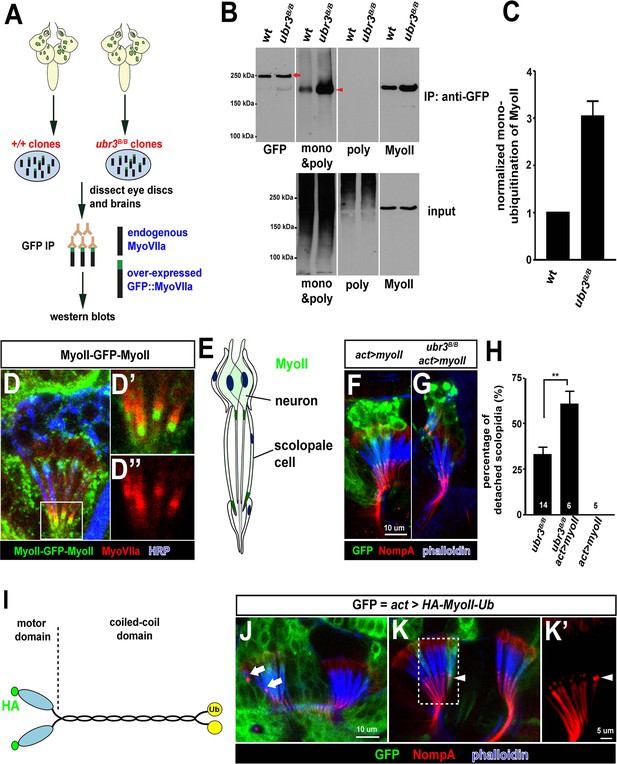
Ubr3 negatively regulates the mono-ubiquitination of MyoII and MyoII-MyoVIIa interaction.
(A) A GFP amino terminal tagged MyoVIIa construct, GFP-MyoVIIa, is expressed in wild type (control) or ubr3 mutant clones in larval eye-antennal discs. The lysate of eye-antenna discs and brains containing ubr3 mutant cells expressing GFP-MyoVIIa protein was immunoprecipitated with GFP nanobody-conjugated beads and examined on western blots. (B) Western blots with anti-GFP, anti-poly & mono-ubiquitin, anti-mono-ubiquitin and anti-MyoII antibodies. (C) Quantification of mono-ubiquitination of MyoII normalized by total amount of MyoII proteins from (B). (D–D’’) Immunolabeling of GFP (green), MyoVIIa (red) and HRP (neurons, in blue) in Johnston’s Organ from a myoII-GFP-myoII transgenic fly. D’ and D’’ show magnified images of the region shown by white box in D. (E) Distribution of MyoII proteins in Johnston’s organ. (F, G) Wild type MyoII over-expressing cells show normal apical structures of scolopidia, whereas ubr3B/B mutant cells expressing wild type MyoII exhibit enhanced detachment of scolopidia. (H) Quantification of detached scolopidia in ubr3B/B mutant cells, ubr3B/B mutant cells over-expressing MyoII and wild type cells over-expressing MyoII. Error bars show SEM. Numbers of flies quantified are shown in the columns. (I) Diagram shows HA-MyoII-Ub in which MyoII is fused to a Ub coding sequence on the carboxyl terminal. (J–K’) Johnston’s organ containing HA-MyoII-Ub expressing clones (labeled by GFP, green) is immunolabeled by anti-NompA (red) and phalloidin (actin, in blue). Arrow marks detached scolopidia. (K–K’) One MyoII-Ub expressing scolopidium exhibits accumulated NompA at the tips, but stays attached to the cuticle from the third segment (arrowheads). This may be a defective scolopidium just before detaching, suggesting that NompA mis-localization happens prior to apical detachment, as opposed to being a consequence of detachment.
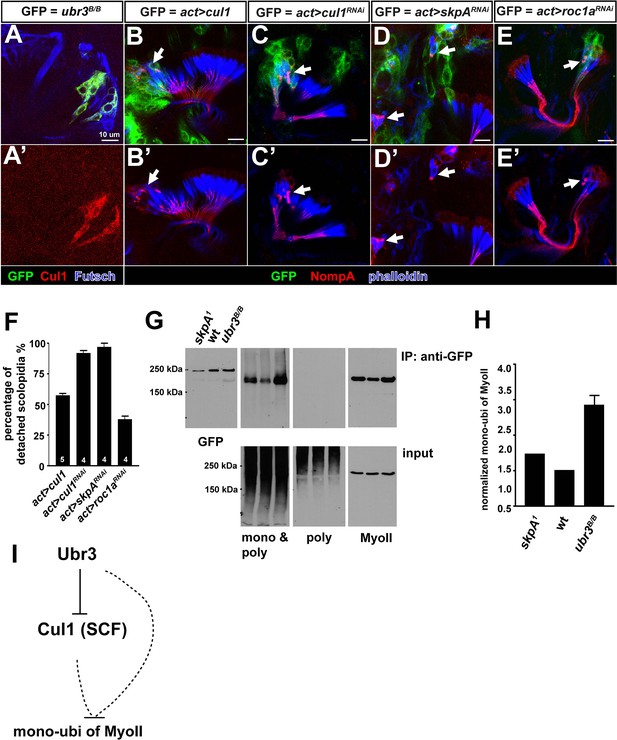
Ubr3 regulates MyoVIIa through Cul1.
(A, A’) Loss of ubr3 (GFP) causes a strong up-regulation of Cul1 in auditory sensory neurons (blue) in Johnston’s organ. (B, B’) Clones over-expressing Cul1 produce similar apical detachment of scolopidia as that observed in ubr3 mutant cells. (C, C’) Down-regulation of Cul1 through RNAi causes apical detachment of scolopidial cells. (D–E’) Down-regulation of skpA or roc1a, SCF E3 ligase components, leads to scolopidial detachment (arrows). (F) Quantifications of detached scolopidia shown in B–E. Error bars show SEM. Numbers of flies quantified are shown in the columns. (G) An amino terminal tagged MyoVIIa construct, GFP-MyoVIIa, is expressed in clones that are wild type (control), skpA or ubr3 mutant cells in larval eye-antennal discs. Eye-antennal discs and brain tissues were dissected from third instar larvae and homogenized in tube. Lysate from the eye-antennal discs and brains was immunoprecipitated with GFP nanobody conjugated beads. Western blots were performed with anti-GFP, anti-poly & mono-ubiquitin, anti-mono-ubiquitin and anti-MyoII antibodies. (H) Quantification of mono-ubiquitination of MyoII normalized by total amount of MyoII from G. Error bars show SEM. (I) A simple working model shows that Ubr3 negatively regulates mono-ubiquitination of MyoII through Cul1 (SCF).
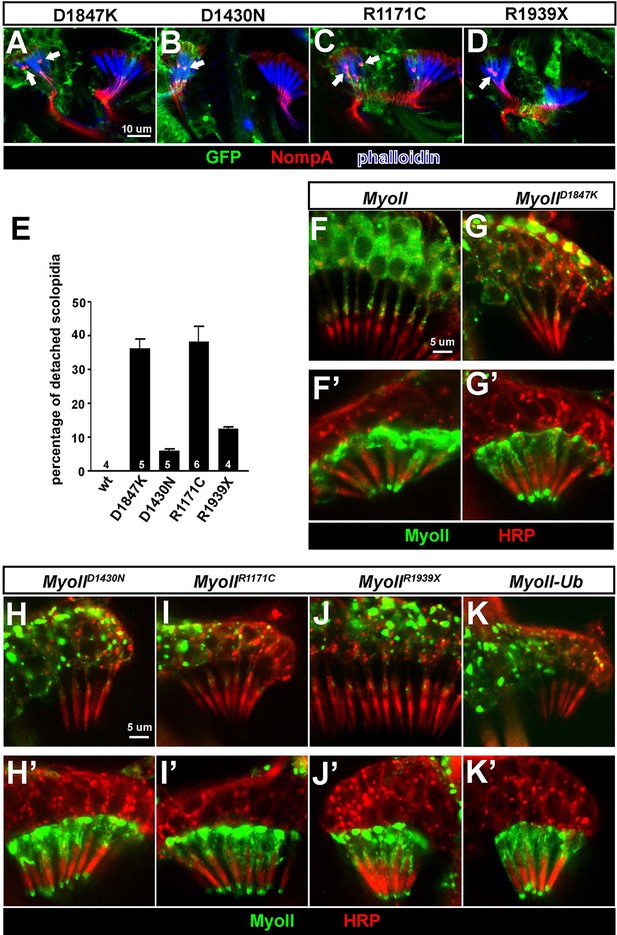
Over-expression of pathogenic variants of MyoII in Johnston’s organ leads to similar defects as ubr3 mutants.
(A–D) Johnston’s organs over-expressing four different GFP tagged MyoIImut in clones (labeled with GFP in green) are immunolabeled with anti-NompA (red) and phalloidin (actin, in blue). Arrows mark detached scolopidia. (E) Quantification of detached scolopidia in the clone cells expressing the four MyoII mutant forms shown in A. Error bars show SEM. Numbers of flies quantified are shown in the columns. (F–K’) Johnston’s organs expressing GFP-MyoII, MyoII mutant forms or MyoII-Ub in neurons using nsyb-Gal4 driver (F–K) or in scolopale cells using nompA-Gal4 (F’–K’) are immunolabeled with anti-GFP (green) and HRP (neurons, in red).
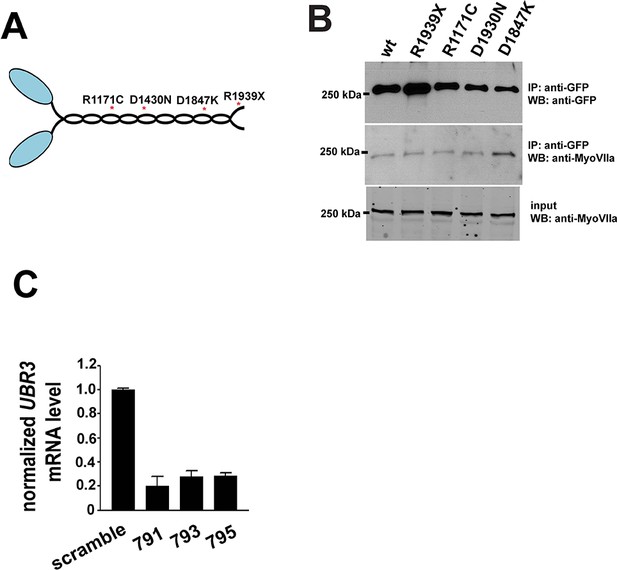
Pathogenic mutations of MyoII.
(A) Structure of MyoII protein and the most common pathogenic mutations used in this study. (B) GFP-tagged wild type MyoII or pathogenic variants of MyoII were expressed in eye-antenna discs using ey-Gal4. Immuno-precipitation was performed using lysate from eye-antenna discs against GFP tags, followed by western blots. (C) RT-PCR results from ARPE-19 cells transfected with different siRNAs.
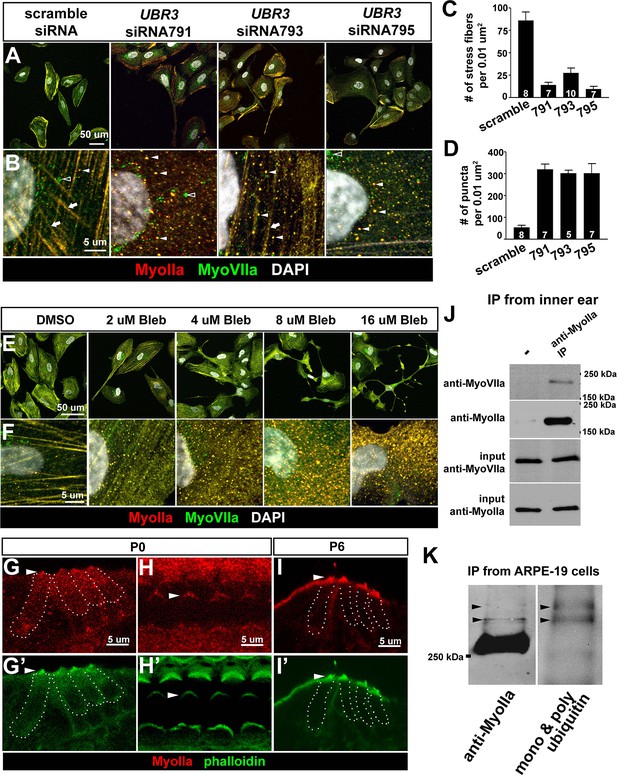
The function of Ubr3 is conserved in vertebrate cells.
(A–B) Cultured ARPE-19 human cells transfected with indicated siRNAs are co-immunolabeled by anti-MyoIIa and anti-MyoVIIa antibodies and DAPI (white). Arrows: stress fibers. Arrowheads: MyoIIa-MyoVIIa co-localized puncta. Empty arrowheads: MyoVIIa positive, MyoIIa negative puncta. (C–D) Quantifications of stress fiber number and MyoIIa-MyoVIIa puncta shown in A–B. Error bars show SEM. Numbers of cells quantified are shown in the columns. (E, F) ARPE-19 cells treated with indicated concentrations of blebbistatin for 30 min followed by immunolabeling with anti-MyoIIa (red) and anti-MyoVIIa (green) antibodies and DAPI. Low concentration (2–4 μM) of blebbistatin treatment resulted in elongated ARPE-19 cells with protrusions, similar as UBR3 siRNA-treated cells. Further increasing the dosage of blebbistatin (8–16 μM) resulted in cells with a more branched, tree-like morphology. The number of puncta correlated with the concentration of blebbistatin, suggesting a specific change in MyoII-MyoVIIa interactions. (G–G’) Immunolabeling of a cochlear section from a neonatal mouse with anti-MyoIIa (red) and phalloidin (green). Hair cells are outlined by dashed lines. (H–H’) Surface view of whole mount cochlea from a neonatal mouse immunolabeled with anti-MyoIIa (red) and phalloidin (green). Arrowheads mark V-shaped stereocilia (labeled by phalloidin, green). (I–I’) Immunolabeling of cochlear section from P6 pup with anti-MyoIIa (red) and phalloidin (green). Arrowheads mark stereocilia (shown by phalloidin staining in green). (J) Co-immunoprecipitation with anti-MyoIIa antibody from P5 cochlear lysate followed by western blotting. (K) MyoIIa was purified from ARPE-19 cells through immuno-precipitation, followed by western blot. Arrowheads indicate ubiquitinated MyoIIa (shown by FK2 antibody).
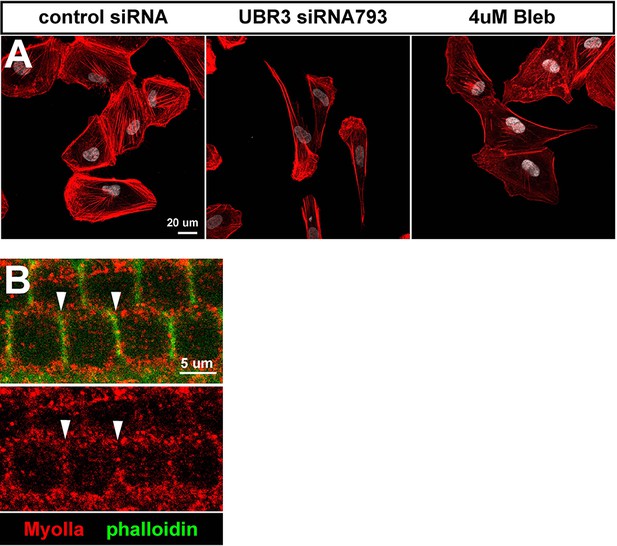
MyoII proteins localize close to cell membrane in the hair cells of mouse cochlea.
(A) Cultured ARPE-19 human cells transfected with indicated siRNAs or treated with 4 μM blebbistatin are co-labled with phalloidin (Actin, red) and DAPI (white). (B) Single section of confocal image shows distribution of MyoIIa (red) in hair cells (labeled by phalloidin, green). MyoIIa proteins localize near cell membrane of hair cells (arrowheads).
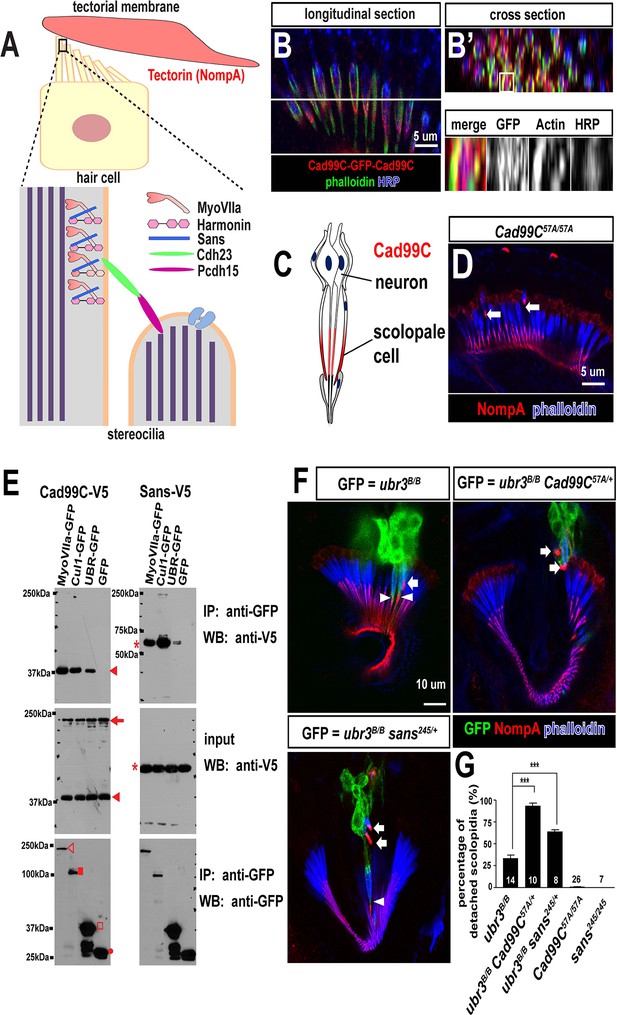
Ubr3, Cul1 and MyoVIIa interact with Drosophila homologues of Usher proteins.
(A) Diagram of a vertebrate hair cell and localization of USH1 proteins in stereocilia. (B–B’) Johnston’s organ of a fly carrying a homozygous GFP knock-in allele of Cad99C is labeled with HRP (blue, neurons), phalloidin (actin, in green, scolopale cells) and anti-GFP (red, Cad99C proteins). (C) Localization of Cad99C proteins in neuronal cilia and the tip region of scolopale cells. (D) Johnston’s organ of Cad99C57A mutant is stained with phalloidin (actin, blue) and NompA (red). Arrows indicate two detached scolopidia. (E) S2 cells transfected with the indicated constructs were lysed and immunoprecipitated with GFP nanobody conjugated beads. Western blots were performed with various antibodies. In the input fraction for the immunoprecipitation, two proteins can be detected: a short 37 kDa carboxyl terminal domain (arrow) and a full length 250 kDa protein (arrowhead). MyoVIIa-GFP (empty arrowhead), Cul1-GFP (square), UBR-GFP (empty square), and untagged GFP proteins (dot). UBR-GFP was used because we could not detect expression of Ubr3 full length protein here. (F) Johnston’s organs containing ubr3B/B, ubr3B/B Cad99C57A/+ or ubr3B/B sans245/+ clone cells (GFP, green) are stained with phalloidin (actin, blue) and NompA (red). Arrows mark detached GFP+ scolopidia and arrowheads mark un-detached GFP+ scolopidia. (G) Quantification of detached scolopidia. Error bars show SEM. Numbers of flies quantified are shown in the columns. ***p<0.001
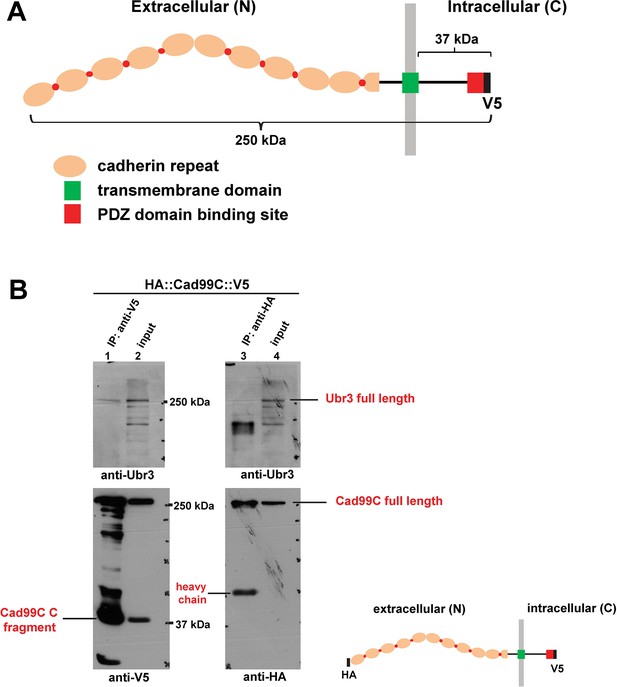
Ubr3, Cul1 and MyoVIIa interact with Drosophila homologues of Usher proteins.
(A) Diagram shows the protein structure of Cad99C. (B) HA-Cad99C-V5 fusion proteins (shown in the right bottom) were expressed in S2 cells. Anti-HA immunoprecipitation or anti-V5 immunoprecipitation was performed with lysate from these cells, followed by western blots with indicated antibodies.
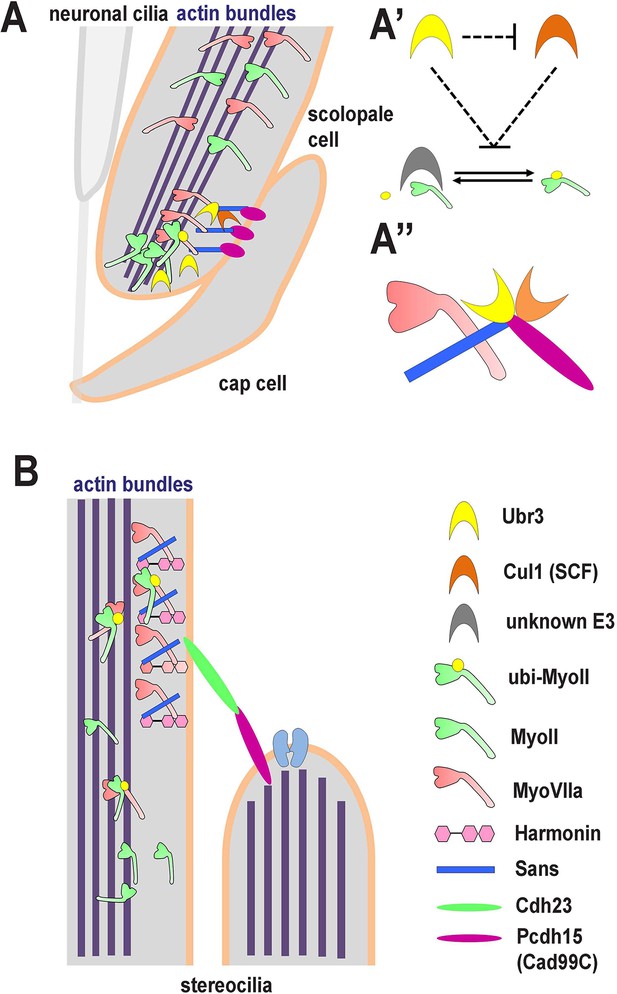
A novel ubiquitination pathway regulates MyoII-MyoVIIa interactions in the auditory sensory organs of Drosophila and mammals.
(A) In Drosophila, MyoVIIa and MyoII are present in the apical regions of scolopidia of Johnston’s organ and are enriched in the tips of the scolopale cells where they contact the cap cell. Ubiquitination of MyoII promotes its interaction with MyoVIIa, the precise level of which is crucial for anchoring the apical junction complexes of the scolopidia. It is possible that the motor activity of either myosin is necessary to transport the complex to the tips of the scolopale cell. Both MyoVIIa and MyoII likely bind to the actin bundles in the scolopale cells and regulate apical attachment of scolopidia. Two Drosophila homologues of Usher syndrome type I proteins, Cad99C (Pcdh15) and Sans, interact with MyoVIIa, Ubr3 and Cul1 in a protein complex. It is not clear whether Cad99C mediates attachment to the cap cells as a homodimer or as a heterodimer with another adhesion molecule. (A’) Ubr3 negatively regulates the level of Cul1 protein. Both Ubr3 and Cul1 inhibit ubiquitination of MyoII indirectly through a pathway involving a third unknown E3 ligase. (A’’) MyoVIIa, Cad99C, Sans, Ubr3 and Cul1 interact as a protein complex. (B) In mammalian hair cells, an USH1 protein complex which includes MyoVIIa, Sans, Harmonin and Cadherin-23 is present close to the stereocilia tips. We speculate that MyoII interacts with MyoVIIa, and that this interaction is promoted by ubiquitination of MyoII. The motor activity of MyoII or MyoVIIa may be required for transport of the MyoVIIa-MyoII-USH1 protein complex to the stereocilia tips.





