SMOC can act as both an antagonist and an expander of BMP signaling
Figures
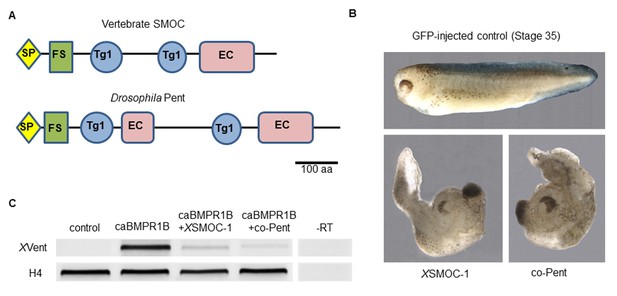
Xenopus SMOC-1 and Drosophila pent are orthologues that inhibit BMP signaling downstream of the BMP receptor.
(A) Schematic representation of vertebrate SMOC and Drosophila Pent: SP- signal peptide, FS – Follistatin-like domain, Tg1 – Thyroglobulin type I-like domain, EC- Extracellular calcium binding domain (B) Dorsalized phenotypes of stage 35 Xenopus embryos following overexpression of mRNAs for XSMOC-1 or codon-optimized pent (co-pent): Xenopus embryos were injected bilaterally at the two-cell stage with 200 pg of mRNA for either GFP (control), XSMOC-1, or co-pent. The exaggerated dorsal/anterior structures and diminished posterior structures observed following XSMOC-1 or co-pent overexpression were observed in 95% of embryos in four independent experiments (n = 130). (C) RT-PCR analysis of animal cap (AC) explants removed from stage 8/9 embryos injected bilaterally at the two-cell stage with either 450 pg of GFP (Control), 150 pg of constitutively active BMP receptor IB (caBMPR-IB) plus GFP (300 pg), or 150 pg of caBMPR-1B plus XSMOC-1 or co-pent (300 pg) mRNAs. The AC explants were incubated until stage 20 before RNA extraction. Induction of the BMP signaling target gene, XVent, by caBMPR-1B was blocked by co-expression of either XSMOC-1 or co-pent. –RT control, without reverse transcriptase.
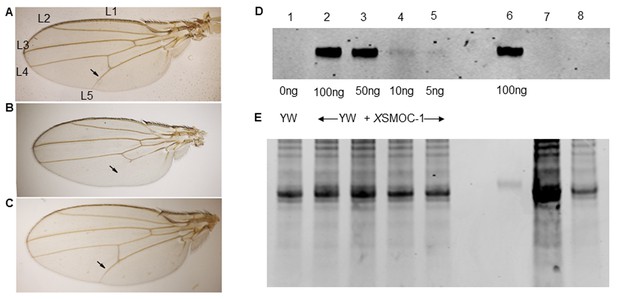
Pent constructs are biologically active in Drosophila, but XSMOC-1 protein is not detected in Drosophila following mRNA overexpression.
(A–C) Adult Drosophila longitudinal wing vein (L1 to L5) patterns in (A) wild type; arrow indicates the position of the anterior crossvein between L4 and L5. (B) pent2-5/pentA17 mutant lacking pent where L5 is missing (C) Rescue of pent2-5/pentA17 phenotype by expression of Pent in a pent2-5/pentA17 background using the UAS/Gal4 system with brinker (brk)/Gal4 as the driver. (D) Immunoblot of Drosophila embryo extracts (10 μg/lane) from control yellow white (YW) Drosophila embryos spiked with XSMOC-1 (Lanes 1–5) as indicated, XSMOC-1 only (Lane 6), or Drosophila lines 4 (Lane 7) and 5 (Lane 8) overexpressing XSMOC-1 mRNA. (E) Coomasie-stained SDS-PAGE gel following protein transfer shown in (D), showing residual protein. The amount of protein in lane seven is notably more than that in other lanes, but the XSMOC-1 signal is nevertheless undetectable.

Nucleotide alignment of pentagone and codon optimized pentagone (co-pentagone).
Amino acid identity is 100%; nucleotide identity is 73%.
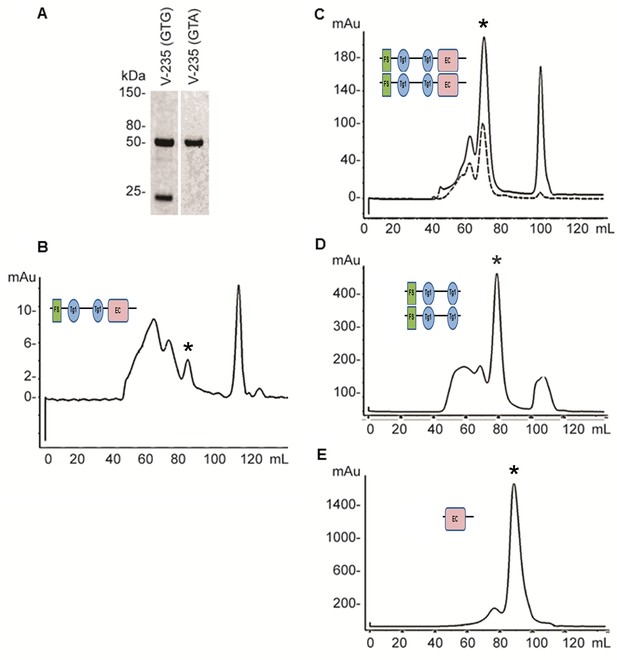
Expression and refolding of recombinant mature XSMOC-1, XSMOC-1∆EC, and XSMOC-1EC.
(A) Coomassie stained SDS-PAGE of wild type (V235-GTG) and silent mutant (V235-GTA) recombinant mature XSMOC-1 following size exclusion chromatography (SEC). The product migrating at 24 kDa is a partial XSMOC-1 sequence beginning at the cryptic start site encoded by GTG at V235. (B–E) Solid lines: SEC profiles obtained following refolding of XSMOC-1 (B, C), XSMOC-1∆EC (D) and XSMOC-1 EC (E) either in the absence (B) or presence of 2 mM Calcium Chloride (C–E). Dashed line: SEC profile (C) obtained for human SMOC-1 refolded in the presence of calcium. Asterisk symbols indicate the peaks corresponding to each schematic diagram.
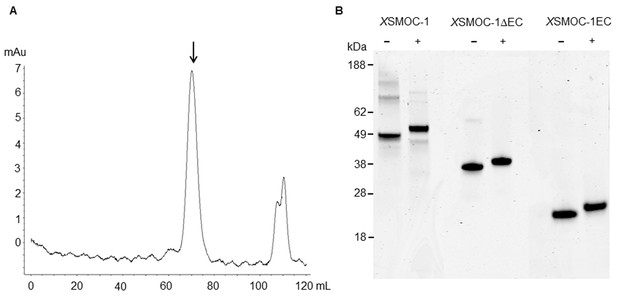
Dimeric XSMOC-1 is not dissociated by chelation or reduction.
(A) SEC profile showing that XSMOC-1, refolded in the presence of 2 mM CaCl2, continues to migrate as a dimer (↓) upon the subsequent chelation of calcium ions by dialysis in the presence of 10.5 mM EDTA. (B) SDS-PAGE analysis of XSMOC-1, XSMOC-1 ∆EC, and XSMOC-1 EC in the presence (+) and absence (−) of β-mercaptoethanol shows that the XSMOC-1, XSMOC-1 ∆EC dimers are not linked by disulfide bonds.
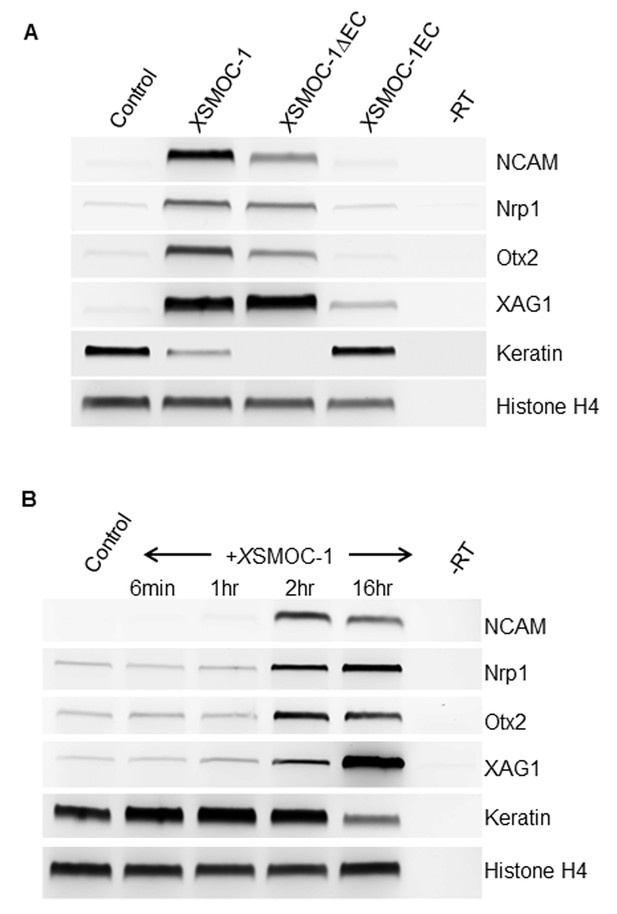
XSMOC-1 and XSMOC-1∆EC, but not XSMOC-1EC convert the fate of naïve Xenopus ectoderm explants (animal caps) to anterior neural tissue within two hours.
(A) RT-PCR analysis of animal caps removed at stage 8/9 and incubated in 0.7X MMR/0.1% BSA (control) containing equimolar amounts of XSMOC-1 (100 μg/ml), XSMOC-1∆EC (75 μg/ml) or XSMOC-1EC (50 μg/ml) until sibling embryos reached the late neurula stage (20); anterior neural markers (N-CAM, Nrp-1, Otx2, Xag-1) were induced by both XSMOC-1 and XSMOC-1∆EC, but not by XSMOC-1EC. Expression of the ectodermal marker Keratin was suppressed by both XSMOC-1 and XSMOC-1∆EC, but not by XSMOC-1EC. (B) Animal caps removed at stage 8/9 were incubated in 0.7X MMR/0.1% BSA (control) in the presence of XSMOC-1 (100 μg/ml) for six minutes, one hour, or two hours before replacing with 0.7X MMR/0.1% BSA and incubating until sibling embryos reached stage 20. RT-PCR analysis shows that a two hour exposure to XSMOC-1was sufficient to induce the naïve ectoderm to express anterior neural markers 16 hr post-pulse; a one hour exposure was not. The continual presence of XSMOC-1 (16 hr) was used as a positive control.
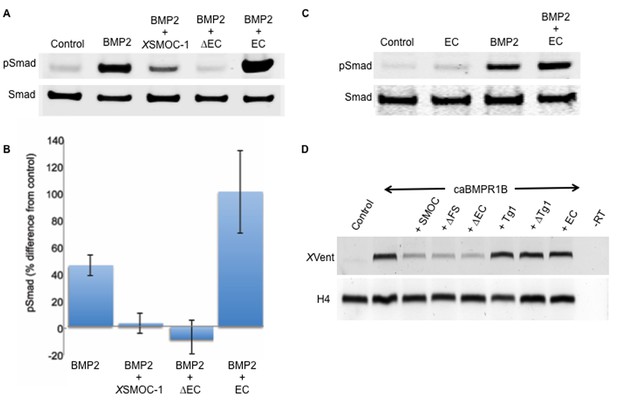
The two thyroglobulin-like domains are necessary for BMP inhibition, whereas the EC domain promotes BMP signaling.
(A) Immunoblot showing phosphorylation of Smad 1/5/8 (pSmad) by BMP2 (50 ng/ml) in HEK-293 cells is inhibited by the addition of XSMOC-1 (100 μg/ml) or XSMOC-1∆EC (75 μg/ml), but is enhanced by the addition of XSMOC-1EC (50 μg/ml). Total Smad is shown as a loading control. (B) Graph showing the relative fluorescence of pSmad 1/5/8 obtained on immunoblots from four separate experiments using both HEK293 and NIH3T3 cells; each treatment is displayed as the percent difference from control. The inhibition of Smad 1/5/8 phosphorylation by XSMOC-1 or XSMOC-1 ∆EC and the potentiation of BMP signaling by XSMOC-1EC are both significant (p=≤0.01). (C) Immunoblot of HEK293 cell extracts showing that XSMOC-1 EC alone does not promote Smad phosphorylation. (D) RT-PCR analysis of Xenopus animal cap (AC) explants removed at stage nine from embryos injected bilaterally at the two-cell stage with mRNAs for GFP (Control), caBMPR1B (120 pg), or caBMPR1B and equimolar amounts of XSMOC-1(600 pg), XSMOC-1 ∆FS (540 pg), XSMOC-1 ∆EC (420 pg), XSMOC-1 Tg1 (330 pg), XSMOC-1 ∆Tg1 (360 pg) orXSMOC-1 EC (240 pg). The AC explants were incubated until stage 20 (late neurula) before RNA extraction and analysis. The induction of the direct BMP signaling target gene, XVent, by caBMPR1B was blocked by co-expression with XSMOC-1, XSMOC-1 ∆FS, or XSMOC-1 ∆EC, but not by XSMOC-1 Tg1, XSMOC-1 ∆Tg1, or XSMOC-1 EC. H4: Histone loading control, –RT: Negative control.
-
Figure 4—source data 1
Source data file for generating Figure 4B.
Absorbance values obtained from pSmad immunoblots in four separate experiments.
- https://doi.org/10.7554/eLife.17935.010
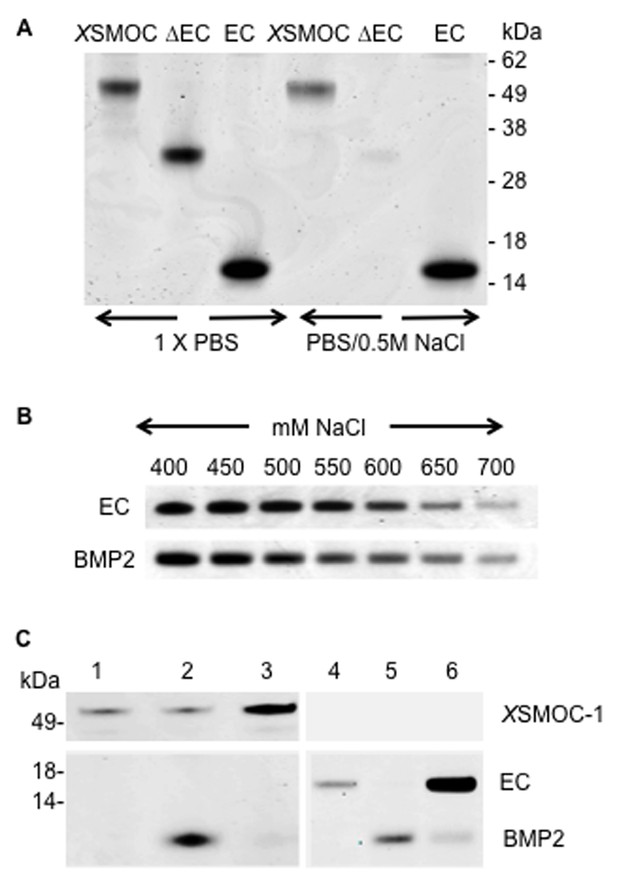
XSMOC-1EC and BMP2 have similar binding affinities for heparin sepharose (HS), but do not bind to each other.
SDS-PAGE analysis (Coomassie staining) of HS elution profiles showing (A) binding of XSMOC-1, XSMOC-1∆EC, and XSMOC-1EC in PBS or PBS/0.5M NaCl; binding of XSMOC-1 to HS requires the EC domain. (B) XSMOC-1EC and mature BMP2 (A284-R396) in a NaCl gradient (400–700 mM) have equivalent HS binding affinities. (C) BMP2 does not bind directly to XSMOC-1 (lanes 1–3) or XSMOC-1EC (lanes 4–6) at physiological ionic strength (PBS); BMP2 (4 μg) incubated with HS (0.3 μl) saturated with XSMOC-1 or XSMOC-1EC (6 μg), did not co-elute with XSMOC-1 or XSMOC-1EC (lanes 3 and 6) and was only present in the unbound fraction (lanes 2 and 5). Saturation of HS by XSMOC-1 and XSMOC-1EC was confirmed by their presence in the unbound fractions (lanes 1 and 4) prior to incubation with BMP2.
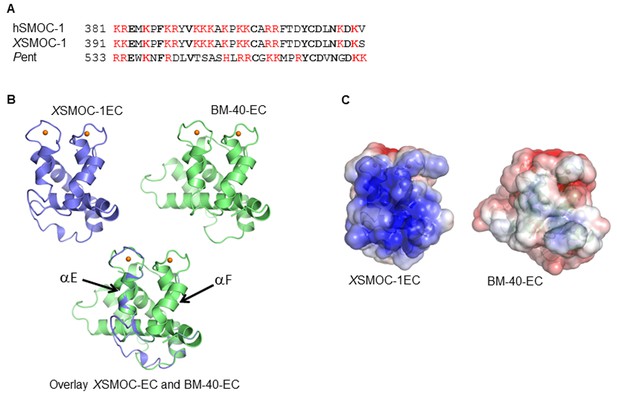
The EC domains of SMOC and Pent are conserved and share structural homology to BM-40.
(A) Predicted heparin binding sequence of hSMOC-1 aligned with the same region of XSMOC-1 and Pent. Conserved amino acids are shown in bold and basic amino acids are shown in red. (B) Unsupervised homology model for the XSMOC-1 EC domain, based on the structure of the EC domain of BM-40 (1SRA.pdb), constructed using the Protein Homology/analogy Recognition Engine (PHYRE, RRID:SCR_010270). The XSMOC-1 EC domain aligns in the exact orientation with the BM-40 model; alpha helices E and F are noted. (C) Electrostatic surface potential maps (based on solvent accessibility) of the EC domains of XSMOC-1 and BM-40 showing an area of positive charge (shown in blue) in XSMOC-1 that is absent in the equivalent region of BM-40.
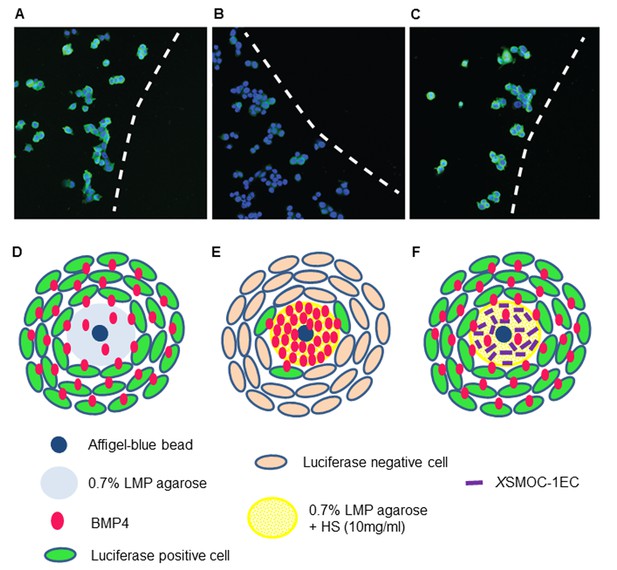
Immunofluorescence assay demonstrating that XSMOC-1 can promote BMP signaling at a distance from its source by competitive binding to HSPGs.
BMP4-soaked beads (100 μm) were implanted into 0.5 μl drops of 0.7% low melting point (LMP) agarose (A, D), LMP agarose containing 10 μg/ml heparan sulfate (B, E), or LMP agarose containing heparan sulfate (10 μg/ml) and 100 μg/ml XSMOC-1EC (C, F) on 8-well chamber slides. C33A-2D2-09 cells, harboring luciferase under the control of a BMP response element (BRE) were seeded at 2 × 104 cells/well and incubated for 48 hr in serum-free medium. Luciferase immunofluorescence (green) indicates cells positive for BMP signaling. Cell nuclei (blue) were stained with DAPI. Dashed lines in A-C indicate the boundaries between C33A-2D2-09 cells and the agarose drops. Analysis of four fields of view in three separate experiments demonstrated the number of luciferase-positive cells to be lower (16% ± 8%) adjacent to matrices containing HS alone (B, E) compared to those containing both HS and XSMOC-1EC (69% ± 26%) (C, F). Representative fields are shown.
-
Figure 6—source data 1
Percentage of luciferase positive cells per field of view.
The number of Luciferase positve cells in five fields of view were calculated as a percentage of the total number of cells in each field.
- https://doi.org/10.7554/eLife.17935.014
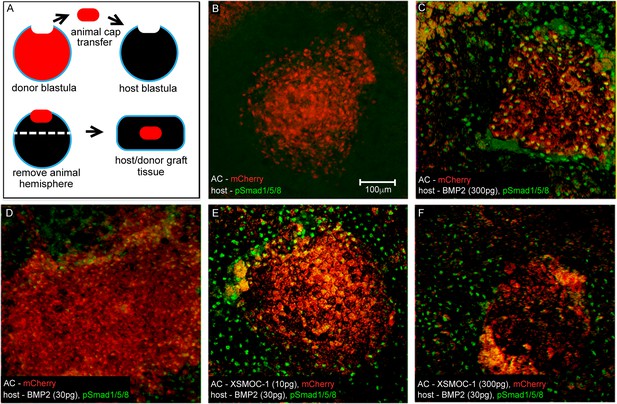
In vivo assay demonstrating that XSMOC-1 can expand the range of BMP2 signaling.
(A) Schematic diagram of the host/donor animal cap (AC) transfer assay. (B–F) Donor AC grafts expressing mCherry (red) and host/donor immunofluorescent nuclear staining of pSmad 1/5/8 (green). (B) Control host + mCherry mRNA (200 pg)-injected donor AC (mCherry AC); endogenous pSmad is not detectable, (C) BMP2 mRNA (300 pg)-injected host + mCherry AC; pSmad is detected throughout the host tissue and donor AC, (D) BMP2 mRNA (30 pg)-injected host (BMP2-30 pg host) + mCherry AC; pSmad is detected in the host tissue and at the host tissue/AC boundary, (E) BMP2-30 pg host + mCherry/XSMOC-1 mRNA (10 pg)-injected AC; pSmad is detected in the host tissue and 4–5 cell diameters into the AC (F) BMP2-30pg host + mCherry/XSMOC-1 mRNA (300 pg)-injected AC; pSmad is not detected in the AC and is also absent at the host tissue/AC boundary.
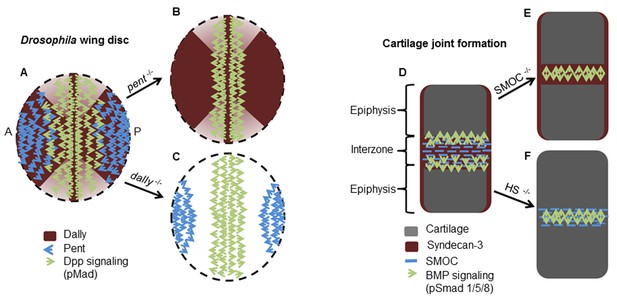
Schematic diagrams showing the proposed mode of action of Pent in the Drosophila wing disc and SMOC in vertebrate joint interzones.
(A) Representations of the expression patterns of pent (Vuilleumier et al., 2010) dally (Fujise et al., 2003) and Dpp signaling (pMad) (Vuilleumier et al., 2010) in wild type Drosophila wing discs, compared with (B) pent−/− or (C) dally−/− mutant discs. The expression patterns observed in the absence of pent or dally (Vuilleumier et al., 2010) are consistent with Pent/Dally binding being required to expand the range of Dpp signaling. (D) Expression patterns of SMOC (Okada et al., 2011; Rainger et al., 2011), Syndecan-3 (Koyama et al., 1995), and BMP signaling (pSmad) [4949] in a developing vertebrate autopod joint, compared with that predicted for (E) the absence of HSPGs (HS−/−), or (F) the absence of SMOC (SMOC−/−).





