A Dynamic molecular basis for malfunction in disease mutants of p97/VCP
Figures
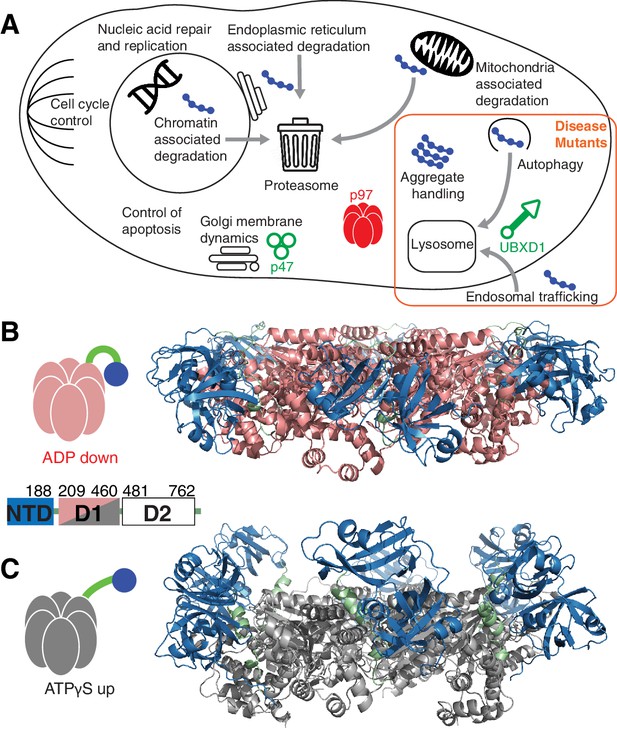
p97 structure and function.
(A) Schematic illustration of a set of p97 cellular functions adapted from (Meyer and Weihl, 2014) highlighting pathways (orange square) that may be affected by IBMPFD disease-related mutations. Substrates are shown in blue and adaptors studied herein in green. (B,C) Ribbon-diagram representation of ND1Lp97 (residues 1–480) [PDB: 1E32 for ADP state, 4KO8 for ATPγS state of R155H] with NTD (blue), linker (green) and D1 (red in ADP state, B; grey in ATPγS state, C) color-coded. Shown to the side are cartoons of ND1Lp97-ADP and ND1Lp97-ATP that are used in other figures, with a single NTD highlighted in the down (ADP) and up (ATP) position, as well as the domain structure of p97.

Impact of nucleotide on the structure of ND1Lp97.
Surface representation of X-ray structures of the ND1Lp97 hexamer (residues 1–480) in ATPγS (A) and ADP (B) states [PDB: 1E32 for ADP state, 4KO8 for ATPγS state of R155H]. Domain organization of full-length p97 is provided in the lower right hand corner of panel (B) and domains are color-coded with NTD in blue, linker in green and D1 in red in the ADP state and grey in the ATPγS-loaded form. Residues that are mutated in IBMPFD patients and that were studied here are highlighted. Note that pairwise contacts between NTD and D1 present in the ADP state (R155H/N387H and R95G/T262A) are lost in the ATPγS state; instead, a new interface forms involving the N-terminus of NTD. Adapted from (Tang et al., 2010).
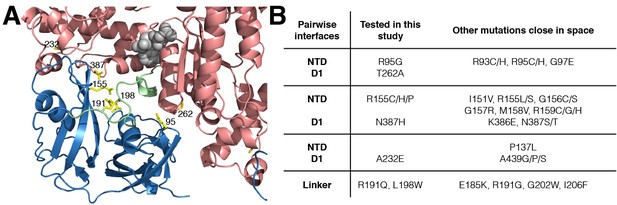
IBMPFD mutation sites.
(A) Residues that are mutated in IBMPFD patients and investigated in this study are indicated in yellow and ADP (grey) is shown with a space-filling model. (B) Table summarizing the distribution of 20 IBMPFD disease mutation sites across three NTD/D1 interfaces and the linker.
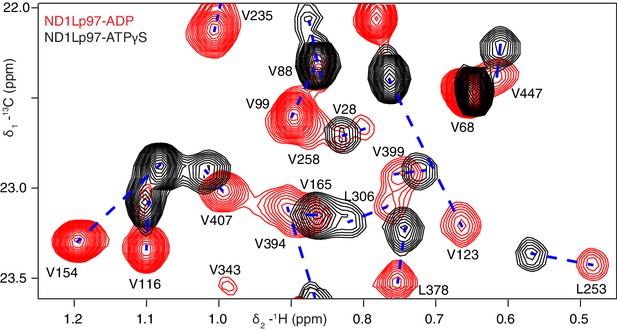
Nucleotide-induced structural changes in p97 as established by NMR.
Superposition of a representative region from methyl-TROSY HMQC spectra of ILVM-13CH3-ND1Lp97 recorded at 800 MHz, 50°C. Dashed lines illustrate peak movement between nucleotide states. Selected methyl assignments have been included.

ND1Lp97 is a good structural model for NTD and D1 of full-length p97.
Superposition of 13C-1H HMQC spectra of [2H,12C, proR ILVM 13CH3]-labeled wt ND1Lp97-ADP (red) and full-length p97-ADP (black), 800 MHz, 50°C highlighting (A) I, (B) V/L, (C) M correlations. Assignment of methyl groups to the protein primary sequence is provided for representative residues. Notably, the spectrum of ND1Lp97-ADP is contained within that of full-length p97-ADP, however a number of cross-peaks for the full-length are reduced in intensity.

Nucleotide-induced ND1Lp97 spectral changes.
Superposition of 13C-1H HMQC spectra of [2H,12C, proR ILVM 13CH3]-labeled wt ND1Lp97-ADP (red contours) and ND1Lp97-ATPγS (black contours), 800 MHz, 50°C highlighting (A) I, (B) V and L and (C) M residues. Assignments are provided for representative methyl groups. Imperfections in the 1H 180° pulse in the center of the HMQC t1 period gives rise to small doublet components (indicated by *) that can be eliminated by replacing the pulse with a composite pulse element.
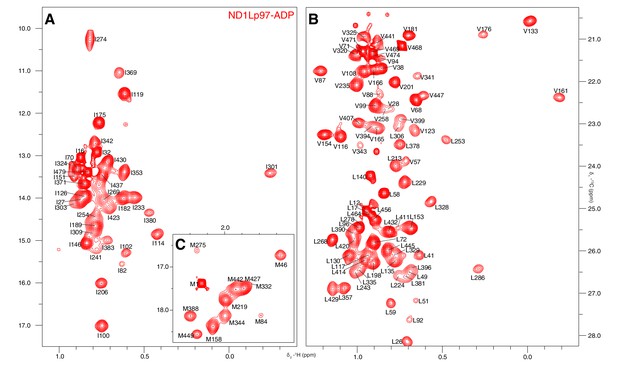
Near complete methyl assignments for ND1Lp97-ADP.
13C-1H correlation map of [2H,12C, proR ILVM 13CH3]-labeled wt ND1Lp97, 800 MHz, 50°C highlighting (A) I, (B) V and L, and (C) M methyl groups.
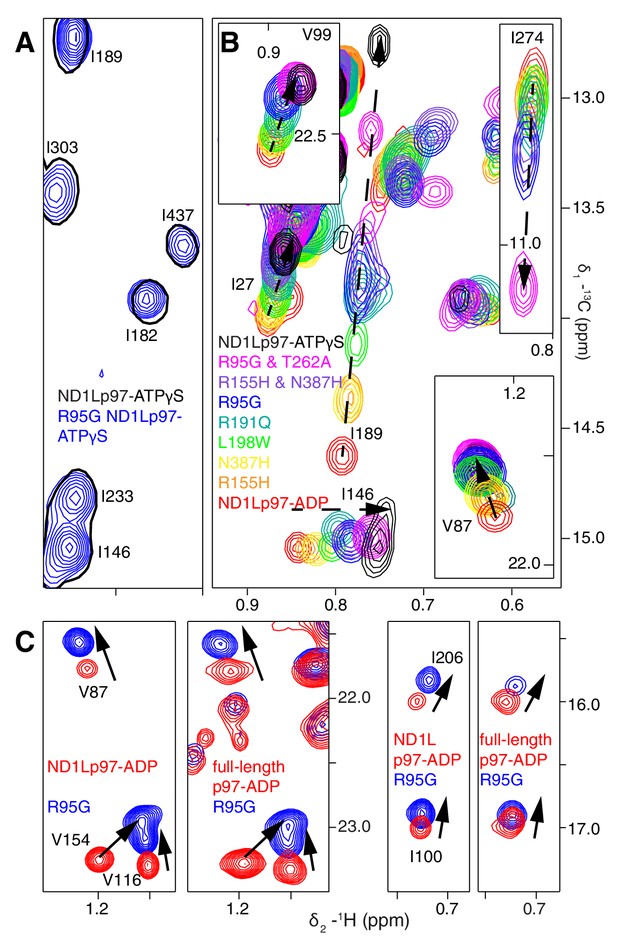
A dynamic NTD equilibrium between up/down states is affected by mutations in ND1Lp97-ADP.
(A) Similar spectra are obtained for wt (black, single contour) and R95G (blue, multiple contours) ND1Lp97-ATPγS. (B) Superposition of selected 13C-1H HMQC spectral regions of wt (red) and mutant ND1Lp97-ADP (colored as indicated) and of wt ND1Lp97-ATPγS (black), focusing on V87, V99, I146 from the NTD, I189 from the NTD-D1 linker and I274 (D1), showing the progressive titration of cross-peak chemical shifts. Note that the I274 peak for wt ND1Lp97-ATPγS is not shown; a large CSP is noted from the ADP to ATPγS substitution due to the proximity of I274 to the nucleotide. (C) Superposition of selected regions of 13C-1H HMQC spectra of full-length wt and R95G p97 (ADP state, right) showing analogous changes as for ND1Lp97-ADP (left).

Disease mutation-induced ND1Lp97 spectral changes.
Superposition of selected regions of 13C-1H HMQC spectra of [2H,12C, proR ILVM 13CH3]-labeled ND1Lp97, 800 MHz, 50°C. (A) Mutations do not change the structure of ND1Lp97-ATPγS. Spectra were recorded on the E305Q Walker B mutant to suppress hydrolysis of the nucleotide (Wendler et al., 2012). (B) Mutations do change the structure of ND1Lp97-ADP. Assignments of methyl groups are provided for representative residues that are affected by mutations in a progressive (essentially linear) manner, as in Figure 4B of the main text. The proximal location of V201 to both L198 and the nucleotide-binding site leads to non-linear CSPs for V201 in the case of L198W and upon binding ATPγS. In analogy to I189, L229, and M388 also have large linear CSPs and pU values have been calculated from CSPs of these residues as well, with very good agreement obtained. For example, pU values of 0.14/0.13/0.13 (R155H mutant), 0.27/0.21/0.21 (L198W), 0.39/0.27/0.33 (R191Q) and 0.42/0.56/0.45 (R95G) are calculated from the CSP trajectories of I189/L229/M388, respectively. In addition to the down state one can prepare a ‘locked-down state’ by generating a cross-link between residues 155 and 387 (see Figure 6D–E), where some but not all of the residues that are sensitive to the up/down equilibrium show further CSPs.
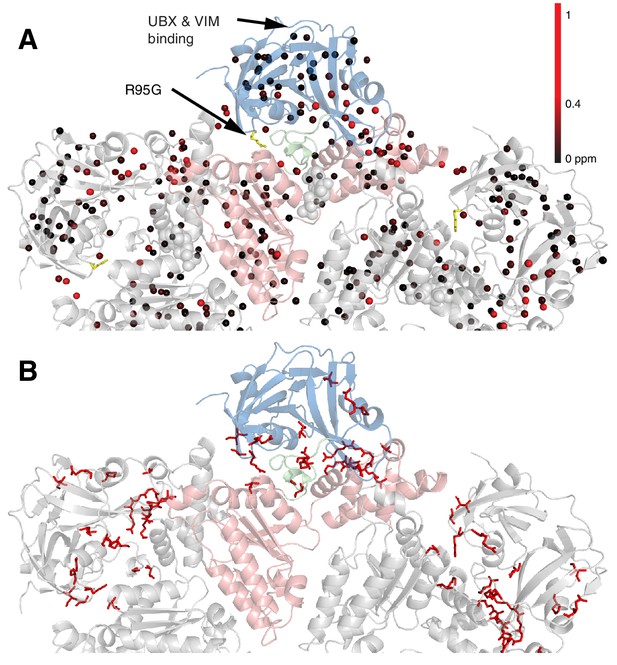
Methyl CSPs introduced by the R95G mutation.
(A) Methyl groups are shown as spheres and color-coded according to the size of the CSP, as indicated. The UBX/VIM (Buchberger et al., 2001; Stapf et al., 2011) binding groove between the two NTD subdomains is unaffected by disease mutations. (B) For reference, the locations of the 22 identified sites of IBMPFD mutations in p97 ND1L are shown in red.
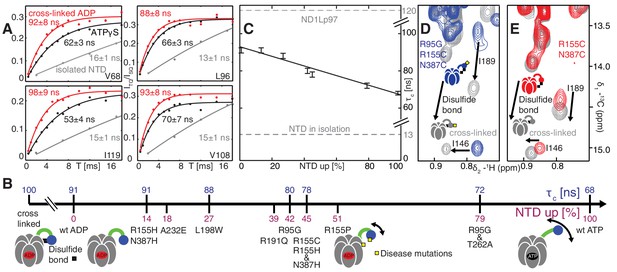
Probing NTD dynamics in ND1Lp97-ADP.
(A) Measurement of NTD tumbling times for ND1Lp97-ADP cross-linked to the down position via a disulfide link between positions 155 (NTD) and 387 (D1) (320 kDa, 50°C, red), ND1Lp97-ATPγS (NTD up, 50°C, black) and isolated NTD (24 kDa, 37°C, grey). Correlation times were obtained via an approach that monitors the build-up of triple quantum coherence (see Materials and methods). (B) NTD tumbling times () and fraction NTD up (), quantified by the 13C chemical shift of I189 for different mutants and nucleotide states of ND1Lp97. Increased motion corresponds to lower . (C) Linear correlation of % NTD up vs NTD for different disease mutants. values for the isolated NTD (adjusted for 50°C) and ND1Lp97 (50°C), calculated from peaks in D1 (see Materials and methods), are given by dashed grey lines for reference. Chemical crosslinking via disulfide bond formation between cysteine residues at positions 155 and 387 (21) forces NTDs of both R95G ND1Lp97-ADP (D) and ND1Lp97-ADP (E, wt at position 95) to the down position, and increases to 100 ns in both cases.
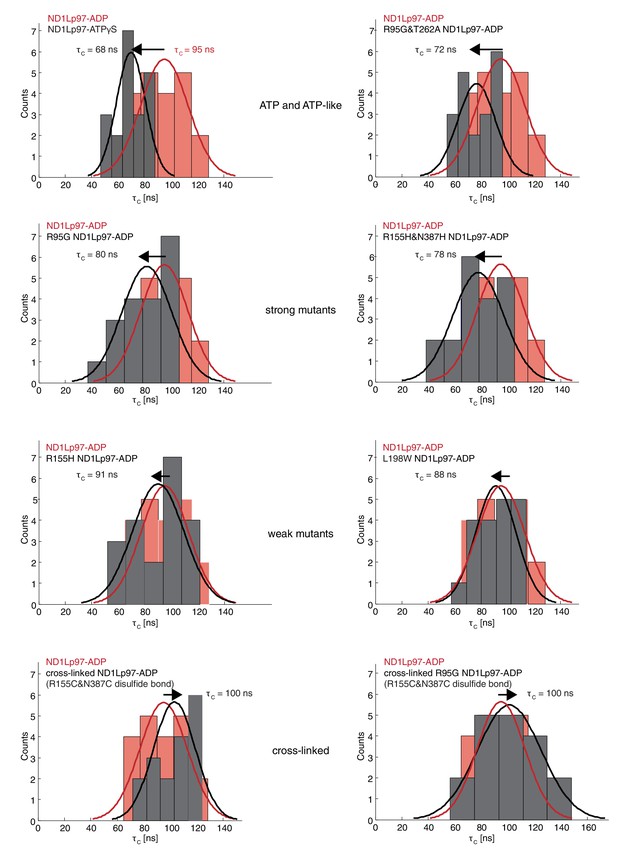
Histograms of τc distributions as obtained from per-residue fits of methyl 1H spin relaxation data.
The distribution for wt ND1Lp97-ADP is shown in red in each panel along with the distribution obtained for each of a series of mutations (dark grey) and wt ND1Lp97-ATPγS that vary the up/down equilibrium from all up (ATPγS) or nearly all up (‘ATP-like’, R95G and T262A) to ~1:1 up/down (‘strong mutants’, R95G and R155H and N387H) to mostly down (‘weak mutants’, R155H and L198W) to locked down (‘cross-linked’ at positions 155 – 387, wt or R95G). A Gaussian fit of each distribution is shown. The indicated values were obtained from a global fit as described in Materials and methods. The direction of the arrow, located above each histogram, illustrates how each perturbation affects , relative to wt ND1Lp97-ADP.
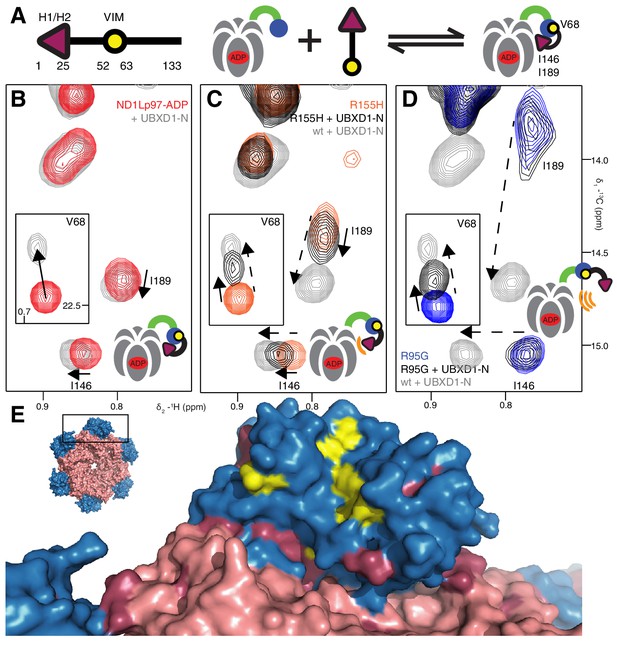
Disease mutants impair binding of UBXD1-N.
(A) (left) Domain organization of UBXD1-N (residues 1- 133 (35)), (right) schematic of binding reaction highlighting 3 key residues that are used as probes of binding in what follows. (B) Selected regions of 13C-1H HMQC spectra of wt ND1Lp97-ADP without (red) and with (grey) 3-fold excess UBXD1-N over protomer, focusing on V68, reporting on VIM domain binding, as well as I146 and I189 that serve as proxies for binding of the H1/H2 motif of UBXD1 (see text). As the NTD is in the down position prior to UBXD1-N binding, only small CPSs are observed for I146/I189, that reflect binding of H1/H2. (C) Addition of 3 fold excess UBXD1-N to R155H results in CSPs (orange to black) that do not extend to the fully bound state observed in wt (grey), indicating only a partial shifting of the up/down equilibrium to the down conformation (cartoon inset). Note that I146/I189 peaks for unbound R155H are shifted upfield relative to wt (compare orange with red peaks) reflecting the increased up population of the NTD for R155H (14%); differences in peak positions for wt and mutant p97 in other panels also reflect changes in the up/down equilibrium. (D) Addition of UBXD1-N to R95G ND1Lp97-ADP (42% up in the unbound state) results in partial binding of VIM but no binding of H1/H2 and subsequently no shift in the up/down equilibrium. (E) Chemical shift perturbations caused by binding of VIM (yellow) and H1/H2 (dark red) of UBXD1 to wt ND1Lp97-ADP mapped onto a surface representation of the NTD (blue) and neighboring D1 structure (inset shows complete hexameric structure from which the surface was taken).
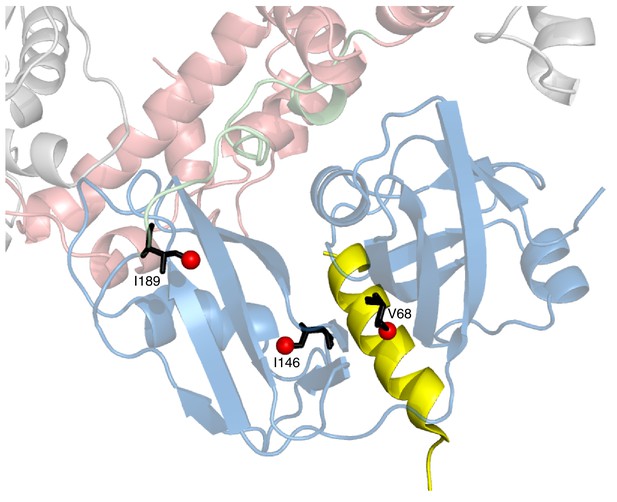
Positions of 3 key reporting residues, V68 (sensitive to VIM binding), I146 and I189 (both Ile are sensitive to the NTD up/down equilibrium) superimposed on a structural model of a complex of the VIM domain (yellow) and ND1Lp97-ADP.
The model has been obtained by aligning the crystal structure of the VIM domain of gp78 bound to p97 NTD (pdb ID 3TIW, [Hänzelmann and Schindelin, 2011]) to the structure of ND1Lp97-ADP (pdb ID 1E32, [Zhang et al., 2000]). Methyl probes are represented as red spheres; note that methyl groups of V68 point directly towards the VIM helix.

Adaptor binding of UBXD1 to wt and mutant p97.
(A) Selected regions of 13C-1H HMQC spectra of [2H,12C, proR ILVM 13CH3]-labeled ND1Lp97-ADP, 800 MHz, 50°C highlighting correlations from V108 and V38 that report on VIM binding (Stapf et al., 2011). In the case of R95G ND1Lp97-ADP the CSPs are smaller than those for wt (compare grey wt with black R95G; 3-fold excess UBXD1-N), since the R95G mutation decreases the macroscopic UBXD1-N binding constant. (B) Selected 13C-1H HMQC spectral regions of wt and R95G ND1Lp97-ADP focusing on residues sensitive to H1/H2 binding. Arrows highlight CSPs that are consistent with a ‘down’ movement of the NTD, which can be derived from the disease mutant series (Figure 4B, Figure 4—figure supplement 1B). Note that the R95G mutant shows no CSPs, indicating no binding to H1/H2, despite the fact that weak binding to VIM occurs. (C–E) As in Figure 7C,D showing further examples of NTD equilibrium shift for weak (C,D) disease mutants (see Figure 7C) and no up/down shift for strong (E) disease mutants (see Figure 7D). M158 (C) and I146, I189 (D,E) that are sensitive to the up/down equilibrium are used as reporters in these cases.

Titration of wt ND1Lp97-ADP with UBXD1-N to obtain an effective macroscopic for the binding reaction.
A value of = 22 ± 2 µM, 50° C, is obtained from a global analysis of chemical shift titration profiles for V68, L140, I175, I182.
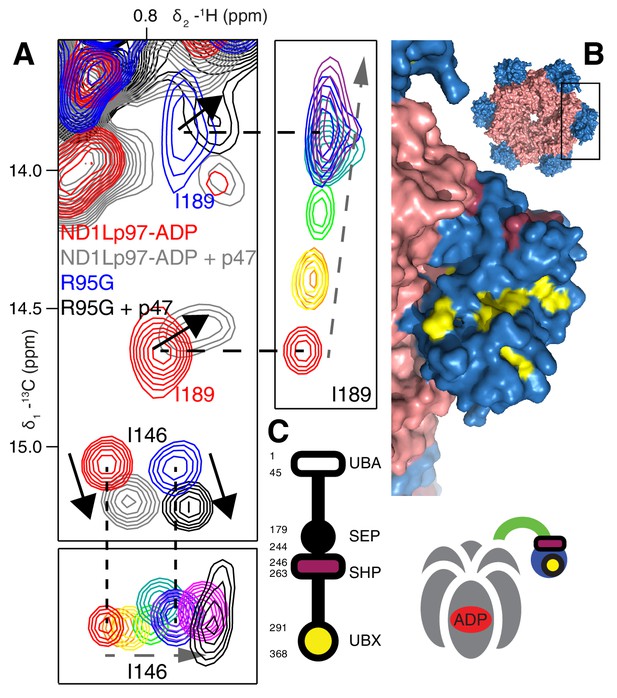
Binding of p47 is not impaired by disease mutations.
(A) Superposition of selected regions of 13C-1H HMQC spectra focusing on I146 and I189 of wt ND1Lp97-ADP and R95G ND1Lp97-ADP without (red for wt, blue for R95G) and with (grey for wt, black for R95G) 1.25 fold excess p47. Similar CSPs indicate that binding to p47 has not been impaired by the R95G mutation, nor is the up/down equilibrium changed. For reference, the ‘mutant titration’ from Figure 4B is provided at the sides of the main spectrum for residues I146 and I189, emphasizing that binding of p47 results in changes that are distinct from NTD up/down. (B) CSPs caused by binding of full-length p47 to wt ND1Lp97-ADP as mapped onto a surface representation of the NTD (blue) and neighboring D1 structure (light red), with the inset showing the complete hexameric structure from which the surface was taken. Residues affected by UBX binding are color-coded in yellow and those perturbed by SHP binding indicated in dark red. (C) Domain organization of p47 and cartoon of the p47-p97 complex, highlighting the regions of interaction.
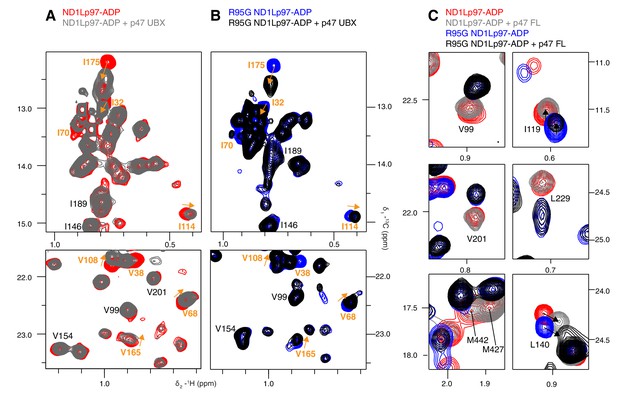
Adaptor binding of p47 to wt and mutant p97.
Selected regions of 13C-1H HMQC spectra of (A) wt and (B) R95G [2H,12C, proR ILVM 13CH3]-labeled ND1Lp97-ADP, 800 MHz, 50°C with and without 1.5 fold excess p47 UBX domain. Assignment of methyl groups is provided for residues involved in UBX binding in orange, as reported in a crystallographic study (Dreveny et al., 2004). Assignments in black indicate residues that respond to the NTD up/down equilibrium shift, which is not affected by binding of the p47 UBX domain. Orange arrows connect peaks that change in position upon binding the UBX domain. (C) Binding of full-length p47 trimer to wt and R95G ND1Lp97-ADP (1.2 fold excess). Residues with CSPs in response to changes to the NTD up/down equilibrium have been selected; note the difference in positions between peaks in red (corresponding to wt ND1Lp97-ADP) and peaks in blue (R95G ND1Lp97-ADP) the reflects differences in pU between wt and R95G p97. Binding of p47 does not alter the up/down equilibrium, as many of the residues that are reporters of the equilibrium (via changes in peak positions upon mutation) do not have CSPs upon p47 adaptor binding (compare red and grey for wt; blue and black for R95G and methyl probes from V99, V201, L229, M427, M442). Alternatively, in the case where binding does cause CSPs (L140, I119), these are not in a direction that indicates changes to the up/down equilibrium.
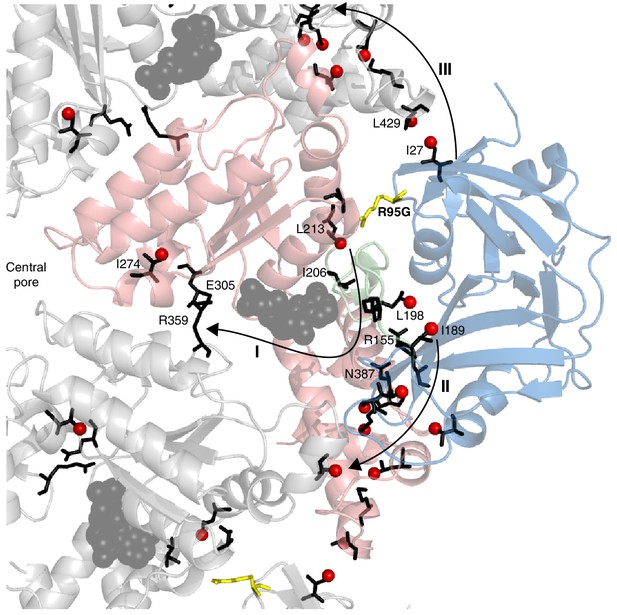
Putative allosteric networks illustrating ‘pathways of communication’ (I-III) from the site of the R95G disease mutation to regions distal in the structure.
R95 is highlighted in yellow. Chemical shift perturbations upon R95G mutation above 0.3 ppm are shown as red spheres. Key residues that form continuous pathways are shown as black sticks, as discussed in the main text.
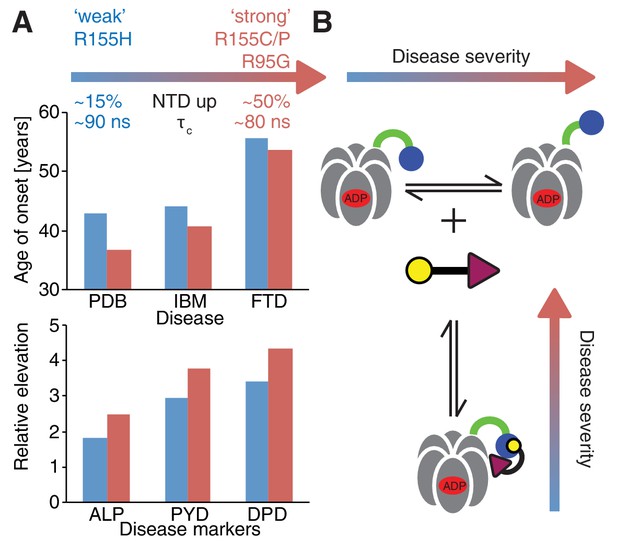
Correlation between disease severity and up/down equilibrium.
(A) Disease (PDB=Paget disease of bone, IBM=inclusion body myopathy, FTD=frontotemporal dementia) onset and elevation of biochemical markers relative to normal (ALP=alkaline phosphatase, PYD=pyridinoline, DPD=deoxypyridinoline) (Mehta et al., 2013), is correlated with the extent of perturbation of the NTD up/down equilibrium. Data from R155H (R155C/P, R95G) patients are used for ‘weak’ (‘strong’) mutants; R155C is associated with a significantly earlier onset of symptoms compared to R155H and showed significantly reduced mean survival (Mehta et al., 2013). (B) Schematic of UBXD1-N binding to ND1Lp97-ADP.