RNase H enables efficient repair of R-loop induced DNA damage
Figures
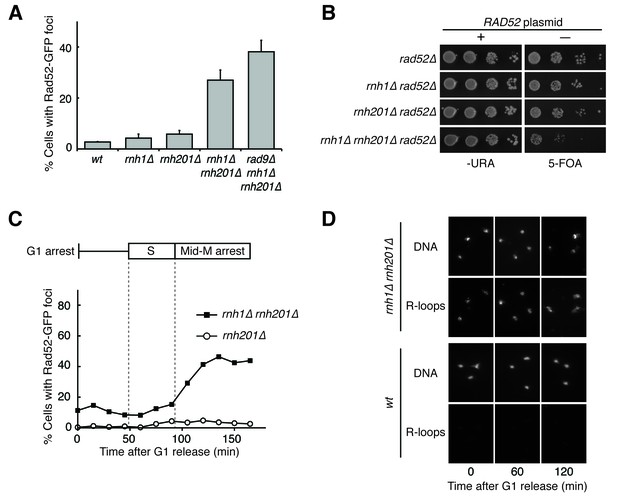
Cells lacking both RNases H accumulate DNA damage in G2-M.
(A) Assessment of Rad52-GFP foci in RNase H mutants. Asynchronously dividing cells were scored for the presence of one or more Rad52-GFP focus. Bars represent mean +/- standard deviation (n = 3; 300 cells scored per replicate). (B) Assessment of Rad52 requirement in RNase H mutants. Cells carrying a plasmid expressing RAD52 and URA3 were plated onto media lacking uracil (-URA, selects for plasmid) or media containing 5-floroorotic acid (5-FOA, selects for plasmid loss). 10-fold serial dilutions are shown. (C) Cell cycle profile of Rad52-GFP foci in RNase H mutants. Synchronously dividing cells were scored for the presence of Rad52-GFP foci. Cells arrested in G1 using alpha factor were washed and released into nocodazole. Samples were taken at 15 min intervals and 300 cells per time point were scored for Rad52-GFP foci. Cell cycle phase is determined by flow cytometry (Figure 1—figure supplement 1). (D) Cell cycle profile of DNA:RNA hybrids in RNase H mutants. Representative images of chromosome spreads of rnh1∆ rnh201∆ and wild-type cells are shown. Spreads are stained for DNA content (DAPI) or immunostained for DNA:RNA hybrids (R-loops) using the S9.6 antibody and a fluorescent-conjugated secondary.
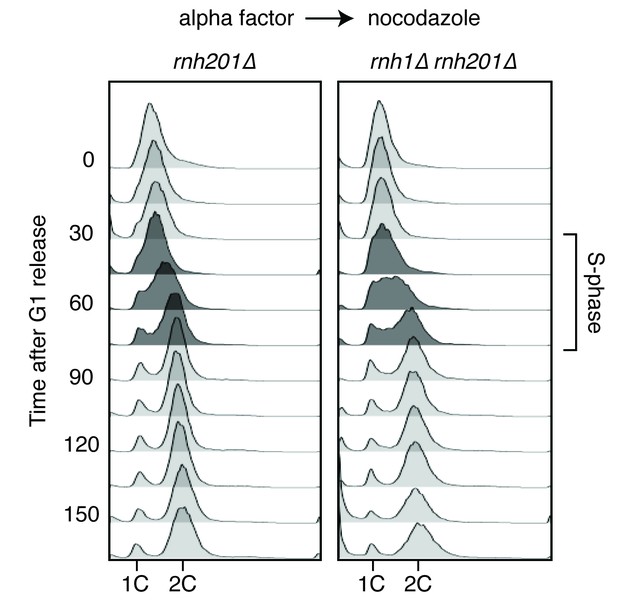
Flow cytometry of rnh201∆ and rnh1∆ rnh201∆ cells released from alpha factor into nocodazole.
Corresponds to Figure 1C.
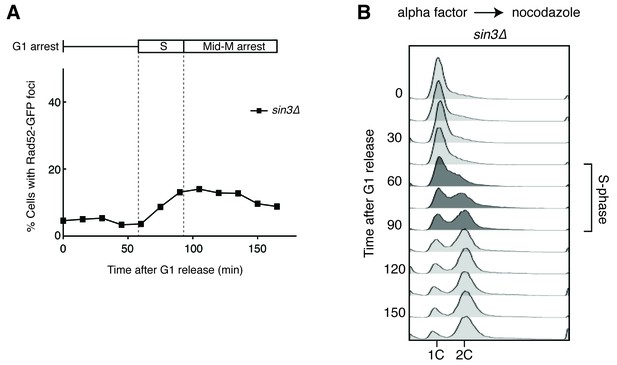
Deleting SIN3 causes increased foci in S phase.
(A) Cultures of sin3∆ cells were arrested in alpha factor and released into nocodazole. Samples were taken at 15 min intervals, and cells were scored for Rad52-GFP foci. (B) Flow cytometry of cells in (A).
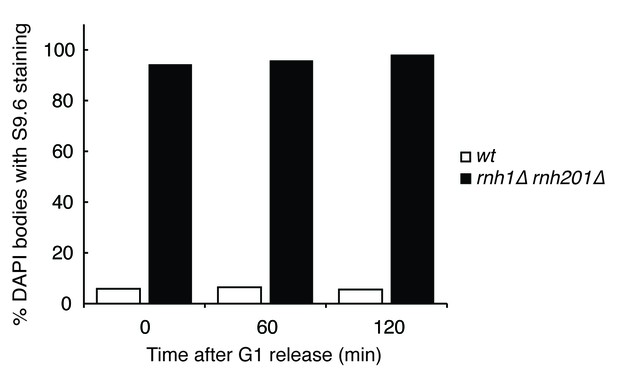
Quantification of Figure 1D.
Chromosome spreads of wild-type and rnh1∆ rnh201∆ cells were scored for either presence or absence of staining with the S9.6 antibody, which appeared uniform across all samples. Bars represent the percentage of DAPI bodies with staining after approximately 300 DAPI bodies were scored.
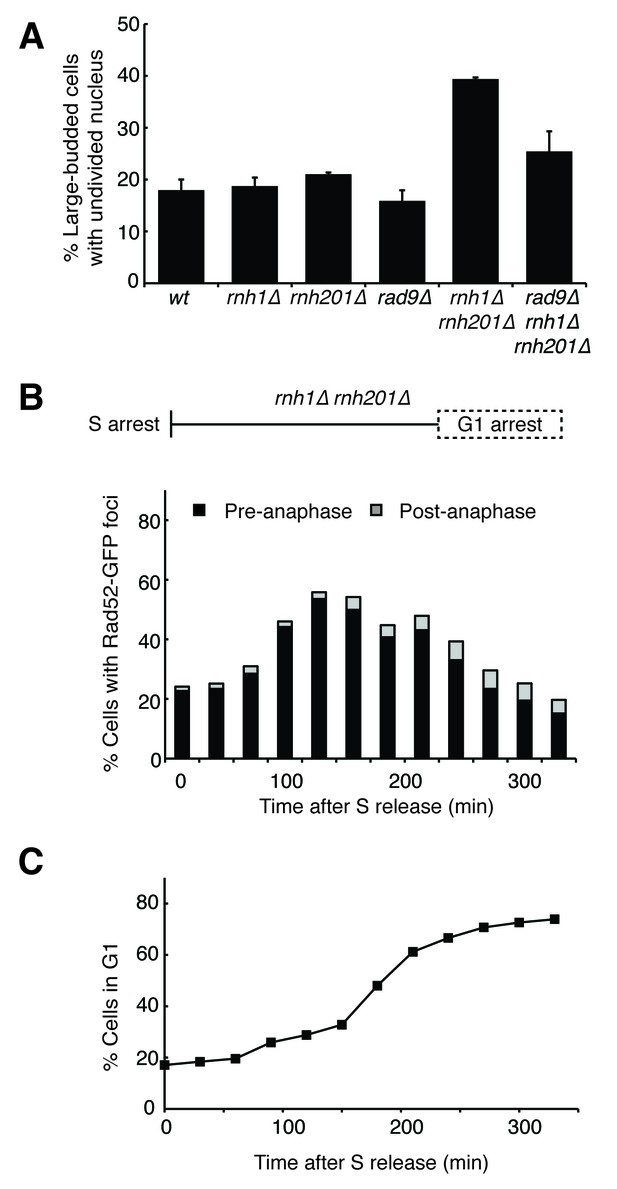
Cells with hybrid-induced DNA damage are slow to repair.
(A) Assessment of cell-cycle delay in RNase H mutants. Asynchronously dividing cells were scored on the basis of their bud size and nuclear morphology. The percentage of cells with large buds and an undivided nucleus (single DAPI mass) are shown. Bars represent mean +/- standard deviation (n = 3, 100 cells scored per replicate) (B) Cell cycle profile of Rad52-GFP foci in dividing cells. rnh1∆ rnh201∆ cells were arrested in S-phase using hydroxyurea, washed, and released into alpha factor. Samples were taken at 30 min intervals and 300 cells per time point were scored for Rad52-GFP foci. If a cell had a Rad52-GFP focus, it was further scored for cell cycle phase. Cells with undivided nuclei (single DAPI mass) are labeled ‘pre-anaphase,’ while those that had undergone nuclear division (two DAPI masses or G1 arrested) are labeled ‘post-anaphase.’ (C) Cell cycle stage of dividing rnh1∆ rnh201∆ cells. Cells from (B) were subjected to flow cytometry (Figure 2—figure supplement 1) and quantified. The percentage of cells with 1C DNA content is shown.
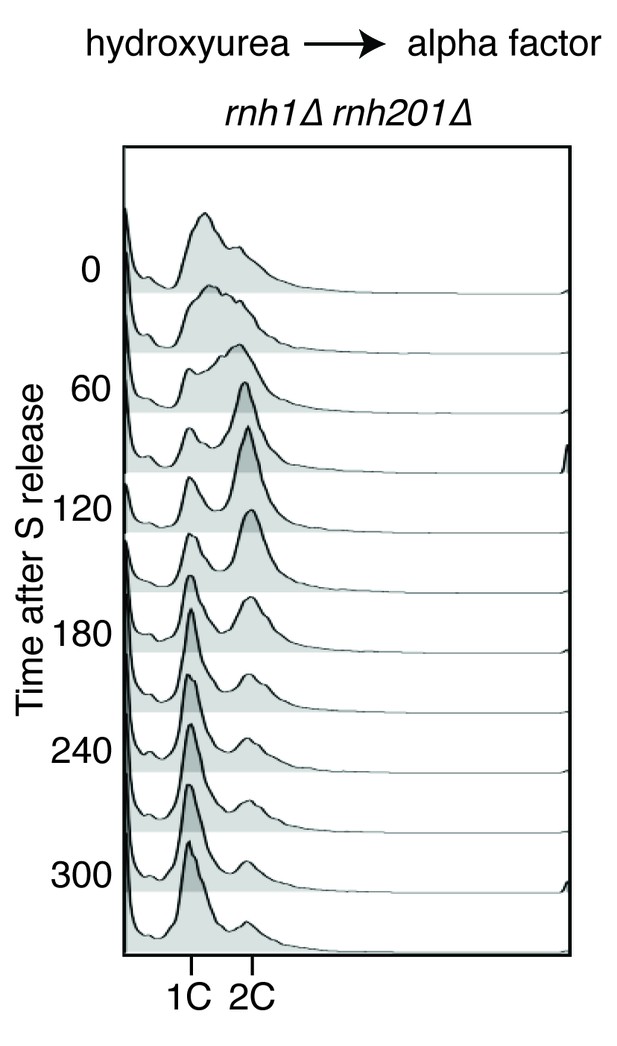
Flow cytometry of rnh1∆ rnh201∆ cells released from hydroxyurea into alpha factor.
Corresponds to Figure 2B.
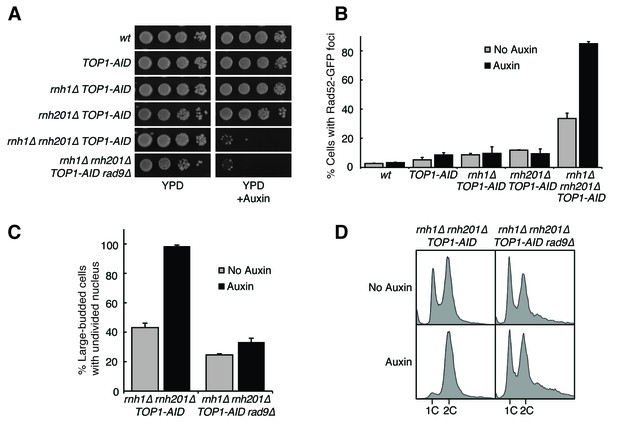
Depleting topoisomerase I exacerbates rnh1∆ rnh201∆ phenotypes.
(A) Assessment of Top1 depletion on viability of RNase H mutants. 10-fold serial dilutions of saturated cultures were plated onto rich media (YPD) or media containing auxin (YPD +Auxin). (B) Assessment of Top1 depletion on Rad52-GFP foci in RNase H mutants. Cultures were grown at 23 degrees and treated with auxin for four hours. Cells were then scored for presence of Rad52-GFP foci. Bars represent mean +/- standard deviation (n = 3, 300 cells scored per replicate). (C) Depleting Top1 leads to robust Rad9-dependent cell cycle arrest. Logarithmically dividing cells were treated with auxin for four hours then scored for bud size and nuclear morphology. The percentage of cells with large buds and undivided nuclei (single DAPI mass) is shown. Bars represent mean +/- standard deviation (n = 3, 100 cells scored per replicate). (D) Cells from (C) were subjected to flow cytometry.
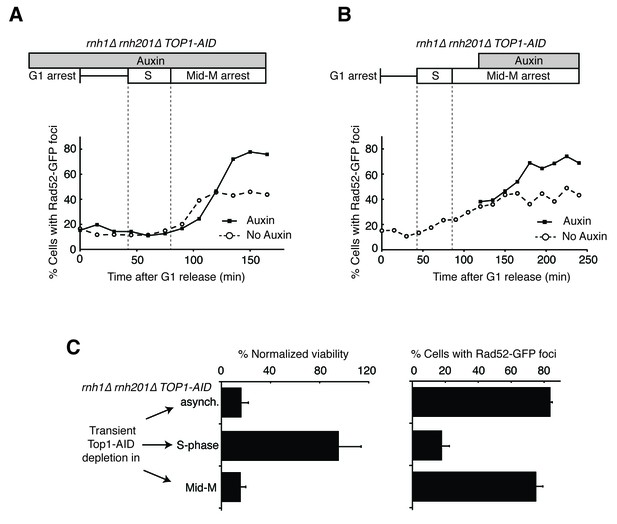
Depleting topoisomerase I causes lethal DNA damage in G2-M.
(A) Depleting Top1 in rnh1∆ rnh201∆ cells shows similar onset of Rad52-GFP at the S/G2-M border. Cultures of rnh1∆ rnh201∆ TOP1-AID cells were arrested in G1 using alpha factor, treated with auxin for 2 hr, then released into media containing nocodazole and auxin. Samples were taken at 15 min intervals and 300 cells per time point were scored for Rad52-GFP foci. (B) Depleting Top1 in rnh1∆ rnh201∆ cells after completion of S-phase causes accumulation of Rad52-GFP foci. Cultures of rnh1∆ rnh201∆ TOP1-AID cells were released from alpha factor into nocodazole. Once cells had completed S-phase, auxin was added. Samples were taken at 30 min intervals and 300 cells per time point were scored for Rad52-GFP foci. (C) Cultures of rnh1∆ rnh201∆ TOP1-AID cells were allowed to divide asynchronously, arrested in S-phase using hydroxyurea, or arrested in Mid-M phase using nocodazole. Once cells were arrested, auxin was added for four hours. Left – Cells were washed and plated on YPD for recovery. Viability was measured by normalizing colony-forming units from auxin-treated cells to untreated cells. Bars represent mean +/- standard deviation (n = 4). Right – Cells were fixed and scored for Rad52-GFP foci. Bars represent mean +/- standard deviation (n = 3, 300 cells scored per replicate).
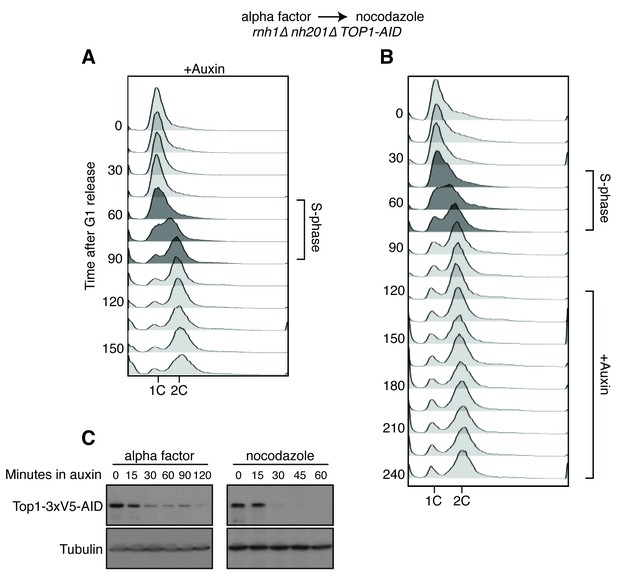
Details on rnh1∆ rnh201∆ TOP1-AID cells released from alpha factor into nocodazole.
(A) Flow cytometry corresponds to Figure 4A. Auxin is added to cells while they are arrested in alpha factor. Cells are then released into nocodazole and auxin, maintaining the state of Top1 depletion. (B) Flow cytometry corresponds to Figure 4B. Auxin is added to cultures in nocodazole 120 min after release from alpha factor. (C) Cells arrested in alpha factor or nocodazole were treated with auxin. Samples were taken at the indicated time points and processed for western blotting. Top: Mouse anti-V5 antibody detects Top1-3xV5-AID. Addition of auxin depletes Top1 by two hours in alpha factor and 30 min in nocodazole. Bottom: Rabbit anti-tubulin antibody detects Tub1.
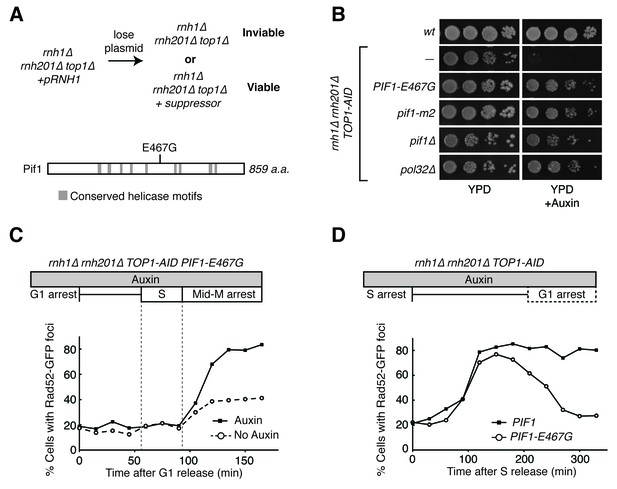
Pif1-E467G allows for repair of R-loop induced damage.
(A) Top: schematic of genetic screen for suppressors of hybrid-induced lethality. Cells for the screen were rnh1∆ rnh201∆ top1∆ and carried a plasmid expressing RNH1 and URA3. Cultures were grown in non-selective media and plated onto 5-FOA to select for cells that had lost the plasmid and therefore gained suppressor mutations of rnh1∆ rnh201∆ top1∆ lethality. Bottom: schematic of Pif1 showing location of E467 relative to evolutionarily conserved SFI helicase motifs and motifs conserved between Pif1 and RecD, as previously published (Boulé and Zakian, 2006). (B) Mutations in PIF1 and POL32 suppress auxin sensitivity of rnh1∆ rnh201∆ TOP1-AID cells. 10-fold serial dilutions of saturated cultures were plated onto YPD or YPD with auxin. (C) Pif1-E467G does not change accumulation of Rad52-GFP foci. Experiment in Figure 4A was repeated on rnh1∆ rnh201∆ TOP1-AID PIF1-E467G cells. (D) Pif1-E467G allows for repair of Rad52-GFP foci. Experiment in Figure 2B was repeated on rnh1∆ rnh201∆ TOP1-AID cells in the presence of auxin with or without PIF1-E467G.
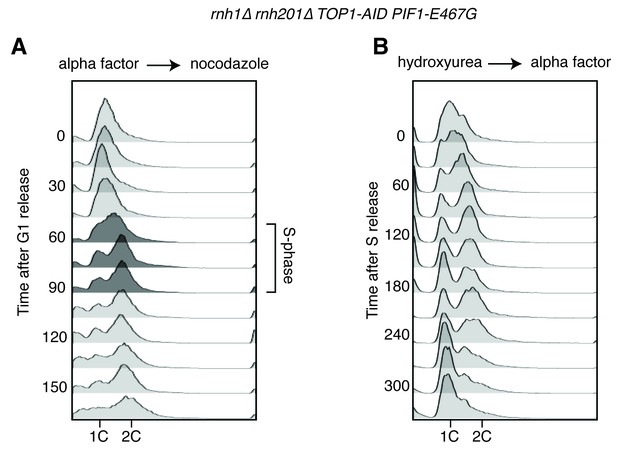
Flow cytometry on rnh1∆ rnh201∆ TOP1-AID PIF1-E467G cells.
All cells have been treated with auxin for two hours before release and auxin is maintained in the culture after release. (A) Cells are released from alpha factor into nocodazole. Flow cytometry profiles correspond to Figure 5C. (B) Cells are released from hydroxyurea into alpha factor. Corresponds to Figure 5D.
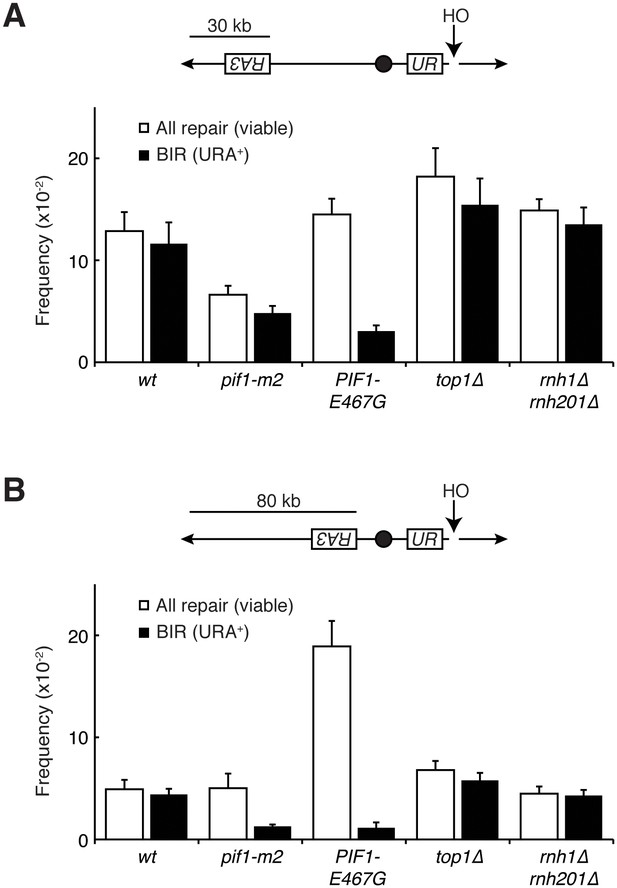
Pif1 mutants inhibit break-induced replication.
(A) Top: Schematic of 30 kb repair template strain. The HO endonuclease is under control of a GAL promoter. In the presence of galactose, it is expressed, inducing a DSB on chromosome V. Sequences telomeric to the HO cut site are non-essential. Homology between the two incomplete URA3 fragments allows for BIR and subsequent telomere addition. Bottom: Frequencies of repair. The percentage of cells that are viable on galactose (compared to total cells plated on YPD) indicates the frequency of all repair events. The subset of those cells that grow on media lacking uracil (URA+) indicates the frequency of BIR events. (B) As in (A), but with a repair template 80 kb from the telomere. Bars represent mean +/- standard deviation (n = 4).
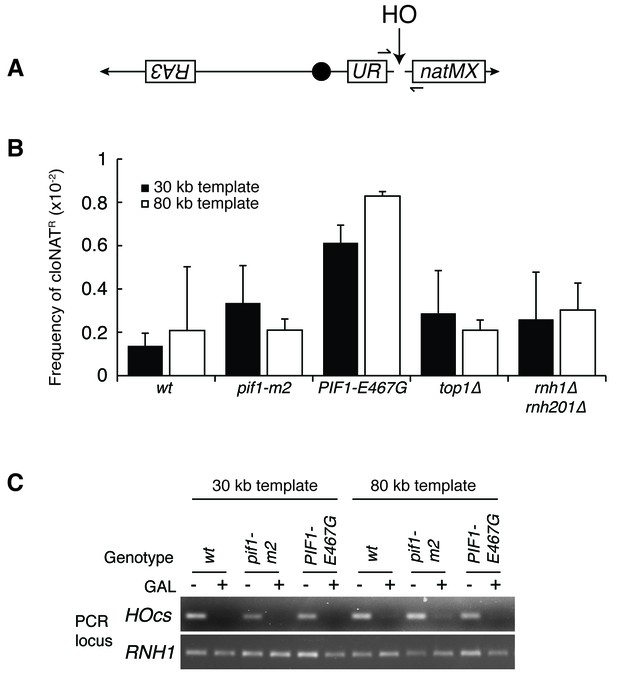
Mutants don’t affect non-homologous end joining or HO-induced DSBs.
(A) More-detailed schematic of chromosome V constructs being used in Figure 6. A gene that confers resistance to the drug cloNAT (natMX) is placed telomeric to the HO cut site (HOcs). Primers designed to flank the HOcs are shown as well. (B) Frequencies of cloNAT resistance in various genotypes in both 30- and 80 kb repair template strains. Viable colonies grown on galactose (Figure 6) were replica-plated to media containing cloNAT. Frequency is calculated by comparing cells that grew on cloNAT to total cells on YPD. Ability to grow on uracil and cloNAT resistance were mutually exclusive. Bars represent mean +/- standard deviation (n = 4). (C) Saturated cultures of the indicated genotype were diluted into media containing dextrose or galactose and allowed to divide for six hours. Genomic DNA was extracted and PCR was performed at the HO cut site (HOcs) or the RNH1 locus as a positive control. PCR products were loaded onto an agarose gel and stained with ethidium bromide.
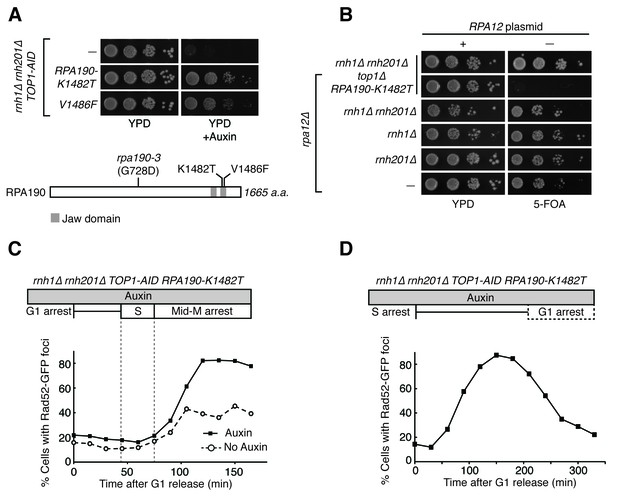
RPA190 mutants allow for repair of R-loop induced damage.
(A) Top: Rpa190-K1482T and -V1486F suppress auxin sensitivity of rnh1∆ rnh201∆ TOP1-AID cells. 10-fold serial dilutions of saturated cultures were plated onto YPD or YPD with auxin. Bottom: Schematic of Rpa190 showing the location of mutations and the jaw domain, as previously published (Engel et al., 2013; Fernández-Tornero et al., 2013). (B) Rpa12 is required in rnh1∆ rnh201∆ TOP1-AID RPA190-K1482T. Cells carrying a plasmid expressing RPA12 and URA3 were plated onto media lacking uracil (-URA, selects for plasmid) or media containing 5-floroorotic acid (5-FOA, selects for plasmid loss). 10-fold serial dilutions are shown. (C) Rpa190-K1482T does not change accumulation of Rad52-GFP foci. Experiment in Figure 4A was repeated on rnh1∆ rnh201∆ TOP1-AID RPA190-K1482T cells. (D) Rpa190-K1482T allows for repair of Rad52-GFP foci. Experiment in Figure 2B was repeated on rnh1∆ rnh201∆ TOP1-AID RPA190-K1482T cells in the presence of auxin.
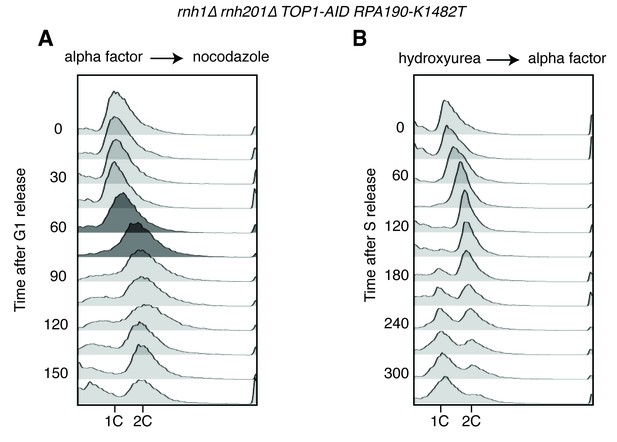
Flow cytometry on rnh1∆ rnh201∆ TOP1-AID RPA190-K1482T cells.
All cells have been treated with auxin for two hours before release and auxin is maintained in the culture after release. (A) Cells are released from alpha factor into nocodazole. Flow cytometry profiles correspond to Figure 6C. (B) Cells are released from hydroxyurea into alpha factor. Corresponds to Figure 6D.
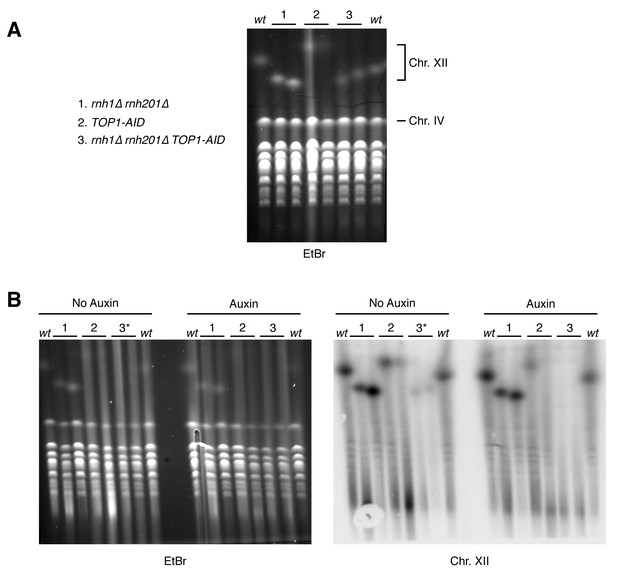
Pulsed-field gel electrophoresis of R-loop mutants.
(A) Two independent isolates each of wild-type (wt), rnh1∆ rnh201∆ (1), TOP1-AID (2), and rnh1∆ rnh201∆ TOP1-AID (3) were grown to saturation in YPD and subjected to PFGE. Gel is stained with ethidium bromide (EtBr). (B) The same cells were diluted, allowed to grow to mid-log phase, and treated with or without auxin. Asterisk indicates rnh1∆ rnh201∆ TOP1-AID cells arrested in nocodazole (3*) to ensure cells are in the same part of the cell cycle as those treated with auxin (G2-M arrested). Left: Gel is stained with ethidium bromide (EtBr). Note that replicating chromosomes from dividing cells run poorly and result in smearing – compare to condensed chromosomes from saturated cultures in (A). Right: Southern blot probing the 35S gene in the rDNA locus on chromosome XII. Note the signal in TOP1-AID (2) and rnh1∆ rnh201∆ TOP1-AID (3) is diminished when cells are treated with Auxin.
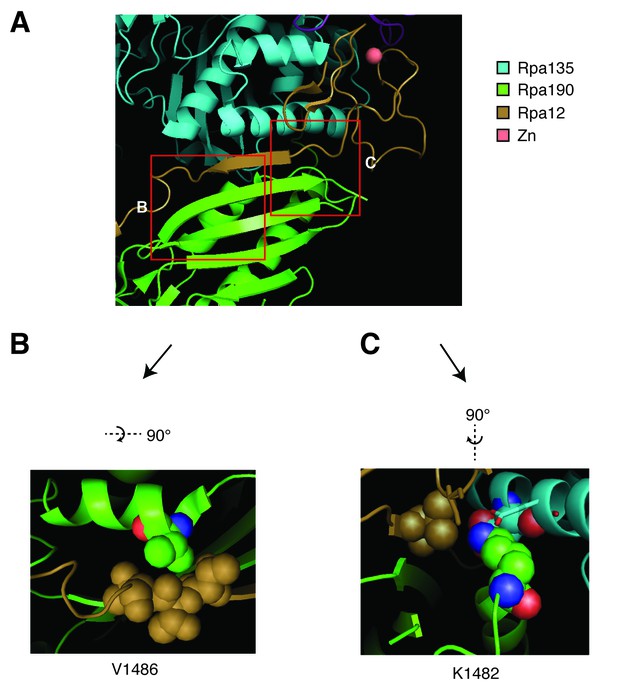
Structural analysis of Rpa190 in the context of the RNA pol I complex.
Structure shown is from Protein Data Bank accession number 4C3I, as originally published in Fernández-Tornero, et al. (2013). (A) The ‘jaw’ domain of Rpa190 (green) is shown with the N-terminal zinc-ribbon of Rpa12 (brown) and the ‘lobe’ domain of Rpa135 (cyan). (B) The highlighted region rotated to see residue V1486 in its position on the alpha helix of RPA190 behind the beta sheet in the foreground of (A). Residue V1486 on Rpa190 is shown in spherical space along with residues T49, T50, and T51 of Rpa12. Potential steric clashes arise between these residues when Rpa190-V1486 is modified to phenylalanine. (C) The highlighted region rotated to see residue K1482 behind the beta sheet in the foreground of (A). Residue K1482 of Rpa190 is shown in spherical space along with Rpa135-D304 and Rpa12-V47 in the background. Also in close proximity are Rpa135-E307 and Rpa12-S6 shown as stick models in the foreground. Modifying Rpa190-K1482 to threonine increases the distance between these residues and possibly interrupts acid-base interactions anchored by the lysine residue.
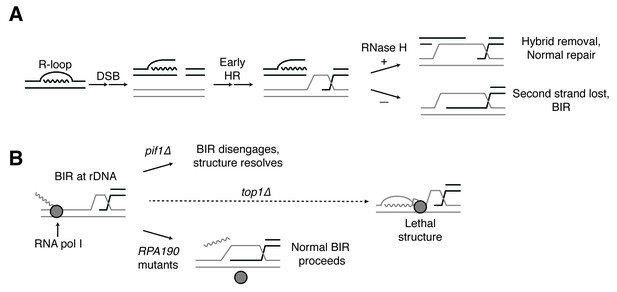
Proposed model for R-loop induced instability.
(A) R-loops that cause DSBs persist at the break site. Early HR events (resection, homology search, strand capture and invasion) proceed as normal for one side of the break, but are inhibited by the presence of an R-loop on the opposite side. If RNase H1 or H2 act to clear the hybrid, repair can proceed as normal. If the hybrid persists, the second strand cannot be captured and the cell engages BIR. (B) BIR at the rDNA encounters replication blocks and slows. These replication blocks (over/under-winding, transcribing or stalled RNA polI, R-loops) are exacerbated in the absence of Top1, creating unresolvable structures that lead to cell death. PIF1 and POL32 mutants make BIR less processive, allowing BIR machinery to disengage before lethality occurs. The repair mechanism used after BIR is disengaged is unknown. RPA190 mutants allow for resolution of these structures, perhaps by disengaging RNA pol I through termination or backtracking activities.
Additional files
-
Supplementary file 1
Strains and plasmids used in this study
- https://doi.org/10.7554/eLife.20533.020
-
Supplementary file 2
Details on candidate mutations in suppressor strains.
Genomes of suppressor strains were sequenced and SNPs were called relative to parental strain JA271a. All non-indel alterations are listed along with the alteration conferred (‘no feature’ – mutation is in an intergenic sequence; ‘syn.’ – synonymous mutation). ‘Primary?’ refers to whether or not this mutation arose in the initial selection step. All primary mutations were then tested for causality. ‘Causative?’ refers to whether or not viable rnh1∆ rnh201∆ top1∆ spores from a cross with the suppressor strain consistently carried the genetic alteration.
- https://doi.org/10.7554/eLife.20533.021





