Nuclear export of misfolded SOD1 mediated by a normally buried NES-like sequence reduces proteotoxicity in the nucleus
Figures
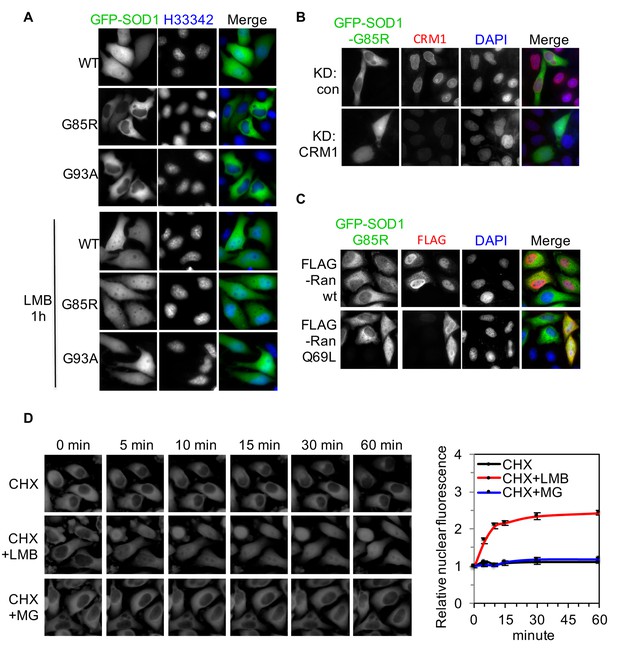
ALS-linked SOD1 mutants are exported from the nucleus by CRM1.
(A) Inhibition of CRM1-dependent nuclear export increases nuclear distribution of SOD1G85R and SOD1G93A. HeLa cells expressing GFP-tagged SOD1wt, SOD1G85R or SOD1G93A were treated with LMB (20 nM) for 1 hr. (B) Knockdown of CRM1 increases nuclear distribution of GFP-SOD1G85R. (C) Overexpression of FLAG-tagged RanQ69L but not wt Ran increases nuclear distribution of GFP-SOD1G85R. (D) Time-lapse imaging of SOD1G85R. HeLa cells expressing GFP-SOD1G85R were treated with cycloheximide (CHX, 100 nM), in combination with MG132 (30 μM) or LMB (20 nM) as indicated. Images were acquired at indicated time points. The nuclear (N) and total (T) GFP fluorescence was measured for each cell with ImageJ. The N/T ratio expressed as mean ± SEM was calculated from 30 cells for each group and plotted. The mean N/T ratios at start point (0 min) were arbitrary set as 1. Paired t-test, CHX vs CHX+LMB: p=0.008; CHX+LMB vs CHX+MG132: p=0.006; CHX vs CHX+MG132: p=0.494.
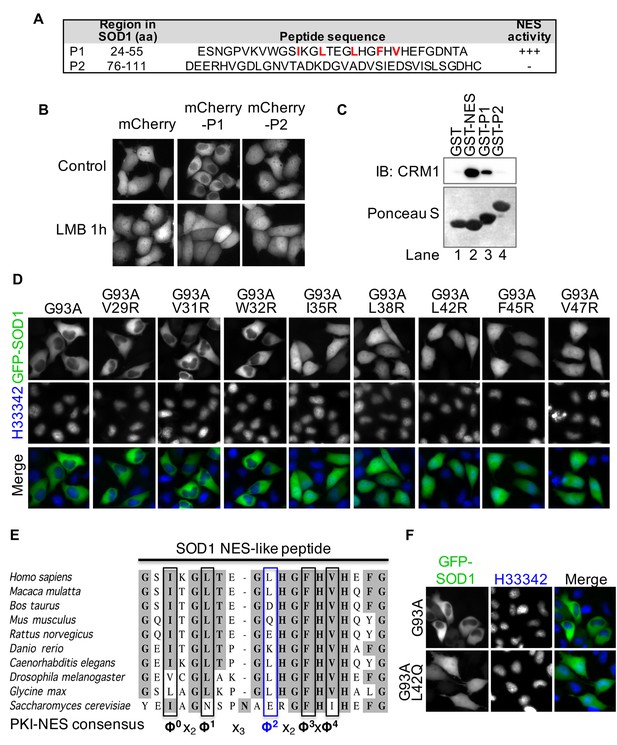
A nuclear export signal (NES)-like sequence is essential for the nuclear export of SOD1 mutants.
(A) Two putative regions containing NES consensus sequences in SOD1 (P1 and P2). (B) P1 or P2 fused with mCherry was expressed in HeLa cells and treated with LMB for 1 hr. (C) GST pull-down. Immobilized GST or GST-tagged PKI-NES (NES), P1 or P2 was incubated with recombinant CRM1 and RanGTP. (D) Identification of the key hydrophobic residues required for nuclear export activity in P1. Each of eight hydrophobic residues in P1 was mutated to Arginine in GFP-SOD1G93A. The mutants were expressed in HeLa cells. (E) Alignment of key residues in P1 with corresponding residues in human SOD1 orthologs and PKI-class NES consensus (Güttler et al., 2010). Note that residue Leu42 in human (Φ2) is not conserved in several other species, including mouse. (F) Mutation of Leu42 in GFP-tagged human SOD1G93A to mouse corresponding residue (Gln) abolishes nuclear export of the mutant SOD1.
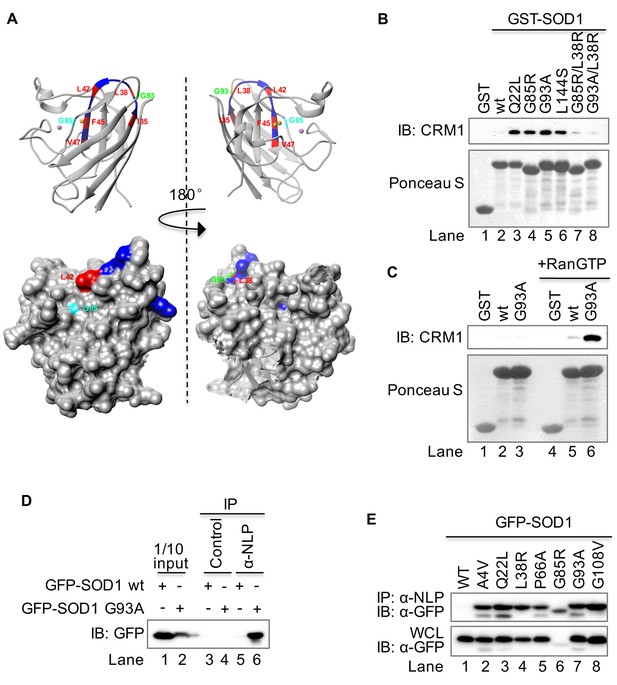
The NES-like sequence is exposed only in SOD1 mutants but not in wt SOD1.
(A) Structural localization of the NES-like peptide in wt SOD1. The X-ray crystallographic structure of wt SOD1 (PDB: 2c9v, only chain A of the dimer was shown) is analyzed in UCSF Chimera 1.8. Key residues in the NES-consensus sequence are labeled in red, whereas other residues are labeled in blue. Note that only Leu42 is exposed in the surface. Upper panel: ribbon structure; lower panel: surface structure. (B) GST pull-down. Immobilized GST or GST-tagged protein as indicated was incubated with recombinant CRM1 and RanGTP. (C) RanGTP-dependent interaction between GST-SOD1G93A and CRM1. Recombinant CRM1 was precipitated by immobilized GST, GST-SOD1wt or GST-SOD1G93A with or without RanGTP. (D) Native immunoprecipitation (IP). GFP-SOD1wt and GFP-SOD1G93A were each expressed in HEK293T cells. The cell lysates prepared under native condition were immunoprecipitated with either the preimmune serum (control) or antibodies against a peptide containing the NES-like sequence of SOD1 (α-NLP). (E) Native IP. HEK293T cells expressing different GFP-tagged SOD1 proteins were lysed under native condition and immunoprecipitated with α-NLP. WCL: whole cell lysates.
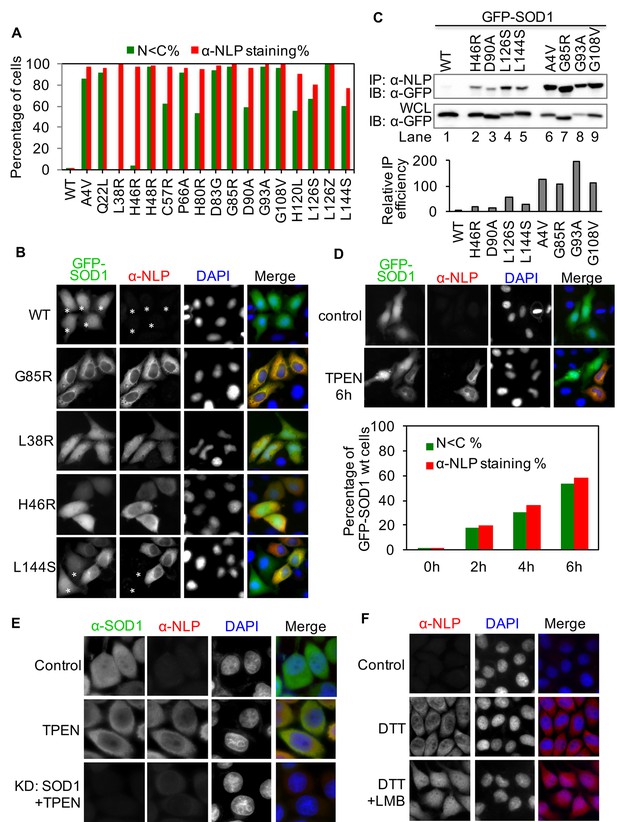
The NES-like sequence is exposed in ALS-linked SOD1 mutants and misfolded wt-SOD1.
(A) HeLa cells expressing GFP-tagged wt-SOD1 or indicated mutants were stained with α-NLP. Percentages of cells exhibiting predominantly cytoplasmic GFP distribution (N<C%) and cells positively stained by α-NLP were plotted. n = 392 (WT), n = 395 (A4V), n = 340 (Q22L), n = 431 (L38R), n = 389 (H46R), n = 411 (H48R), n = 423 (C57R), n = 376 (P66A), n = 393 (H80R), n = 402 (D83G), n = 450 (G85R), n = 435 (D90A), n = 408 (G93A), n = 495 (G108V), n = 355 (H120L), n = 367 (L126S), n = 462 (L126Z), n = 410 (L144S). (B) Anti-NLP specifically stained SOD1 mutants but not wt-SOD1 as detected by immunofluorescence microscopy. Asterisks indicate SOD1-expressing cells not being stained by α-NLP. (C) Native IP. HEK293T cells expressing different GFP-tagged SOD1 proteins were lysed under native conditions and immunoprecipitated with α-NLP. Immunoprecipitates and whole cell lysates (WCL) were blotted with α-GFP antibody. Relative IP efficiency for each SOD1 protein was calculated as the ratio of the band density from IP sample over that from corresponding WCL sample and was plotted as fold-difference relative to wt-SOD1. (D) Demetallation induces exposure of the NES-like sequence in GFP-SOD1wt. HeLa cells expressing GFP-SOD1wt were treated with TPEN (10 μM) and then stained with α-NLP. n = 392 (0h), n = 404 (2h), n = 527 (4h), n = 505 (6h). (E) Control HeLa cells or HeLa cells transfected with SOD1 siRNA (KD: SOD1) were treated with TPEN (4 μM) for 20 hr. Then the cells were stained with α-NLP and α-SOD1 antibodies and imaged for immunofluorescence. (F) DTT induces exposure of the NES-like sequence in endogenous SOD1. HeLa cells were treated with DTT (0.4 mM) and LMB (5 nM) as indicated for 20 hr. Then the cells were stained with α-NLP.
-
Figure 4—source data 1
Data for Figure 4A,C and D.
- https://doi.org/10.7554/eLife.23759.007
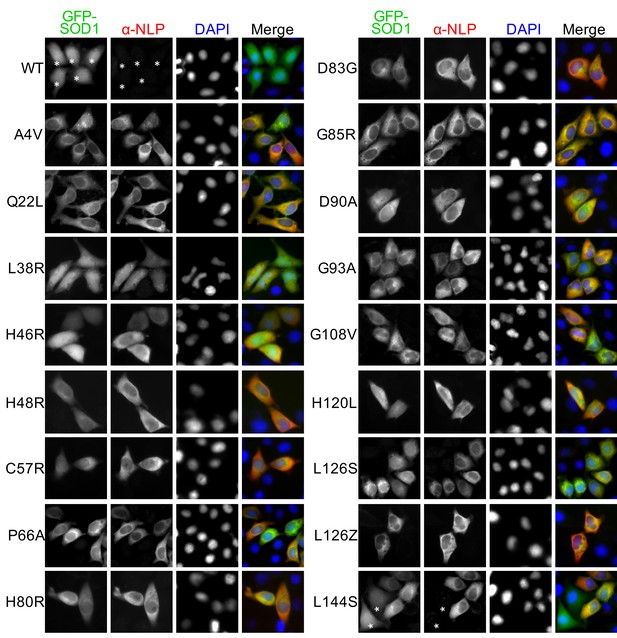
Anti-NLP specifically recognizes various ALS-linked SOD1 mutants but not wt SOD1.
HeLa cells expressing GFP-SOD1wt or indicated mutants were stained with α-NLP. Asterisks indicate GFP-SOD1wt and mutant expressing cells negative for α-NLP staining.
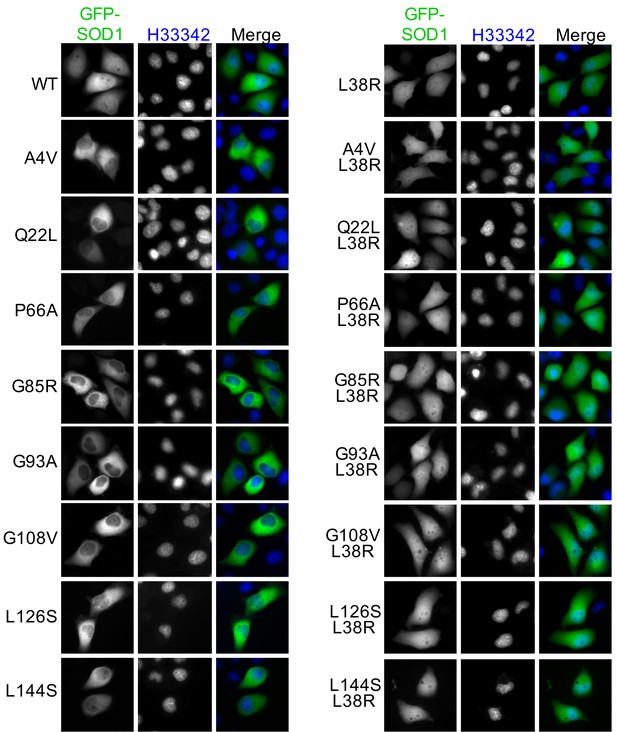
A L38R mutation restores the nuclear distribution of various ALS-linked SOD1 mutants.
GFP-SOD1wt and indicated mutants were expressed in HeLa cells and imaged by fluorescence microscopy.
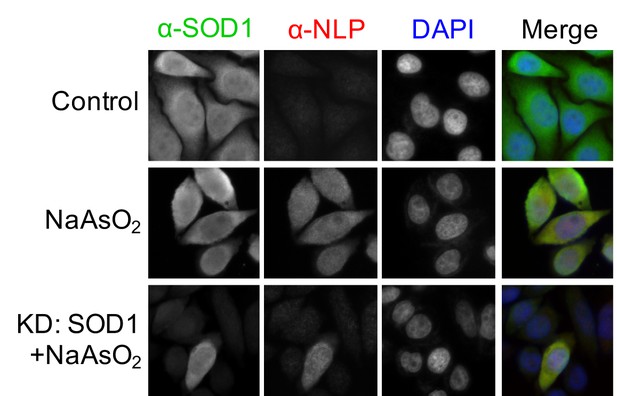
Oxidative stress induces exposure of the NES-like sequence in endogenous SOD1.
Control HeLa cells or HeLa cells transfected with SOD1 siRNA (KD: SOD1) were treated with NaAsO2 (10 μM) for 20 hr. Then, the cells were stained with α-NLP and commercial α-SOD1 antibodies and examined by immunofluorescence microscopy.
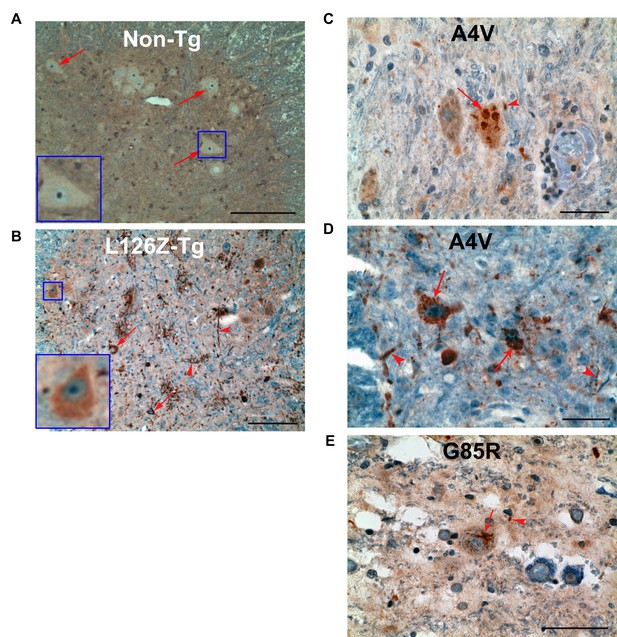
Exposure of the NES-like sequence in spinal cord sections from transgenic mice and ALS cases.
(A). Spinal cord sections from non-transgenic mice were stained with α-NLP. Representative anterior horn neurons are indicated by arrows and also showed in the inset. Scale bar: 100 m. (B) Spinal cord sections from SOD1L126Z transgenic mice were stained with anti-NLP. Representative SOD1 aggregates in anterior horn neurons and neuritis are indicated by arrows and arrowheads, respectively. Inset shows a representative anterior horn neuron with diffused α-NLP staining predominantly in the cytoplasm. Scale bar: 300 m. (C) to (E) Autopsy spinal cord sections from two SOD1A4V ALS patients (C and D) and a SOD1G85R ALS patient (E) were stained with anti-NLP. Representative SOD1 aggregates in anterior horn neurons and neurites are indicated by arrows and arrowheads, respectively. Scale bar: 100 m.
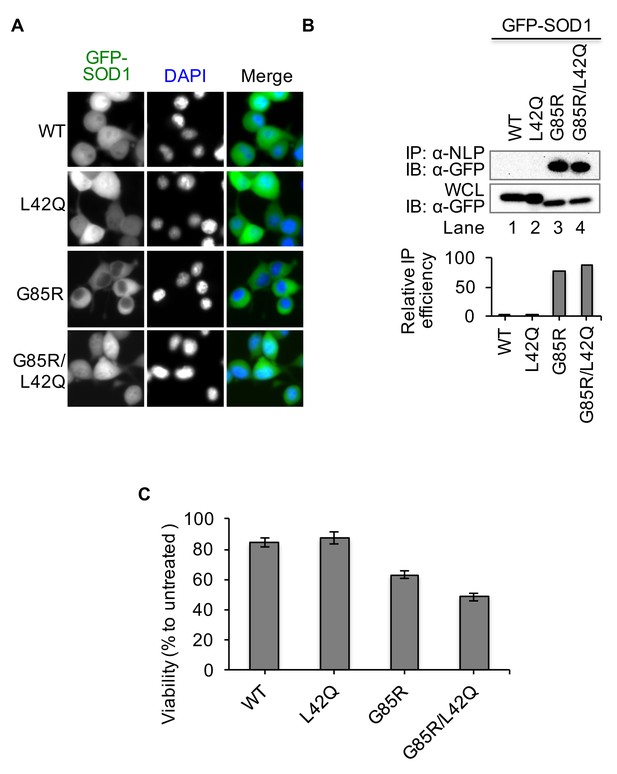
Disruption of the NES by L42Q mutation in SOD1G85R mutant results in higher cytotoxicity in NSC34 cells.
NSC34 cells were infected with lentiviruses expressing GFP-tagged WT, L42Q, G85R or G85R/L42Q SOD1 proteins. (A) SOD1L42Q and GFP-SOD1G85R/L42Q have similar subcellular localizations as GFP-SOD1wt in NSC34 cells. (B) Native IP. NSC34 cells expressing different GFP-tagged SOD1 proteins were lysed under native condition and immunoprecipitated with α-NLP. Immunoprecipitates and whole cell lysates (WCL) were blotted with α-GFP antibody. Relative IP efficiency for each SOD1 protein was calculated as the ratio of the band density from IP sample over that from corresponding WCL sample and was plotted as fold-difference relative to wt-SOD1. (C) Cell viability (WST-1 assay). NSC34 cells expressing GFP-tagged SOD1 proteins as indicated were treated with MG132 (5 μM) for 24 hr. Data represent means ± SEM, n = 4. Unpaired t-test, G85R/L42Q vs G85R: p=0.005; WT vs L42Q: p=0.564.
-
Figure 6—source data 1
Data for Figure 6B and C.
- https://doi.org/10.7554/eLife.23759.013
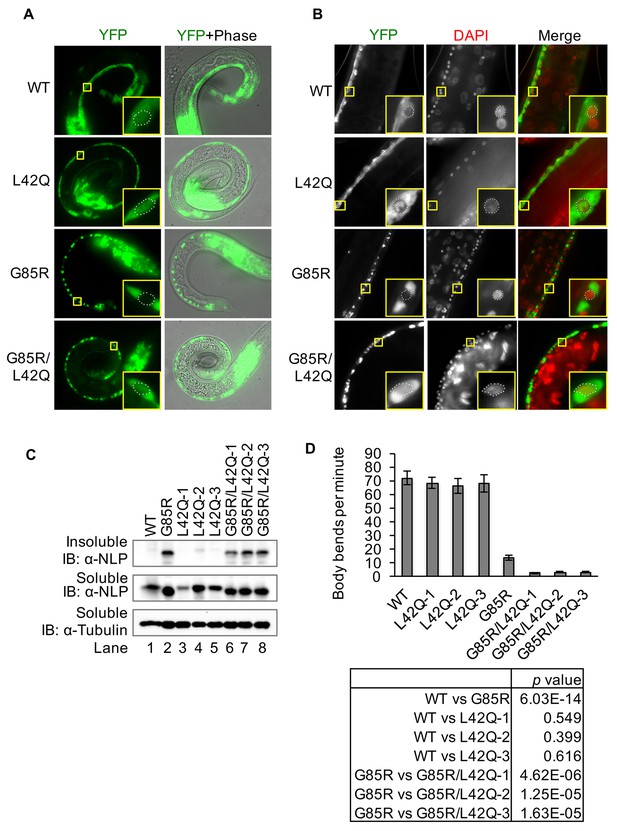
Disruption of the NES by L42Q mutation in SOD1G85R mutant causes severe defects in locomotion in transgenic C. elegans.
Transgenic worm lines neuronal specifically expressing SOD1wt-YFP (WT), SOD1L42Q-YFP (L42Q), SOD1G85R-YFP (G85R) or SOD1G85R/L42Q-YFP (G85R/L42Q) in neurons were generated previously or in this study. (A), Expression of YFP-tagged SOD1 proteins in L1 of transgenic worm lines. Insets show higher magnification of motor neurons. The dot lines depict the nuclear profiles of the enlarged neurons (also in B). (B) Microscopy of adult animals. DAPI (pseudo colored red) was used to stain the nucleus. (C) Immunoblotting. L1 larvae were lysed by sonication on ice. The soluble (S) and insoluble (P) fractions were processed for immunoblotting. (D) Body bending rates. L4 animals were transferred to a drop of M9 buffer and counted for their total body bends in 1 min. Data represent means ±SEM, n = 20. Two-tailed unpaired t-test was used to calculate the p values.
-
Figure 7—source data 1
Data for Figure 7D.
- https://doi.org/10.7554/eLife.23759.015
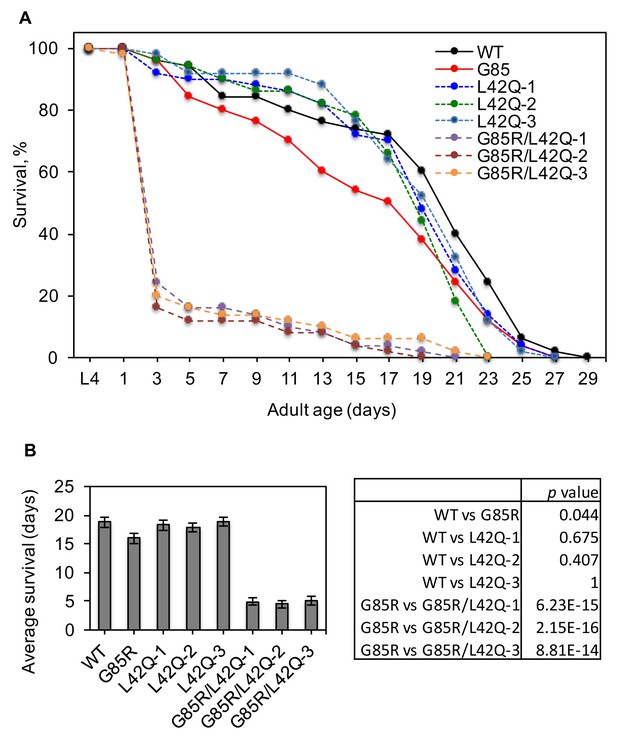
Disruption of the NES by L42Q mutation in SOD1G85R mutant decreases survival of transgenic C. elegans.
(A) Survival curves. Mid-L4 animals were picked and followed for their survival. The surviving worms were transferred to fresh plates every 2 days until all the worms died. n = 50 for each transgenic line. (B) Average survival days from (A). Two-tailed unpaired t-test was used to calculate the p values.
-
Figure 8—source data 1
Data for Figure 8A and B.
- https://doi.org/10.7554/eLife.23759.017
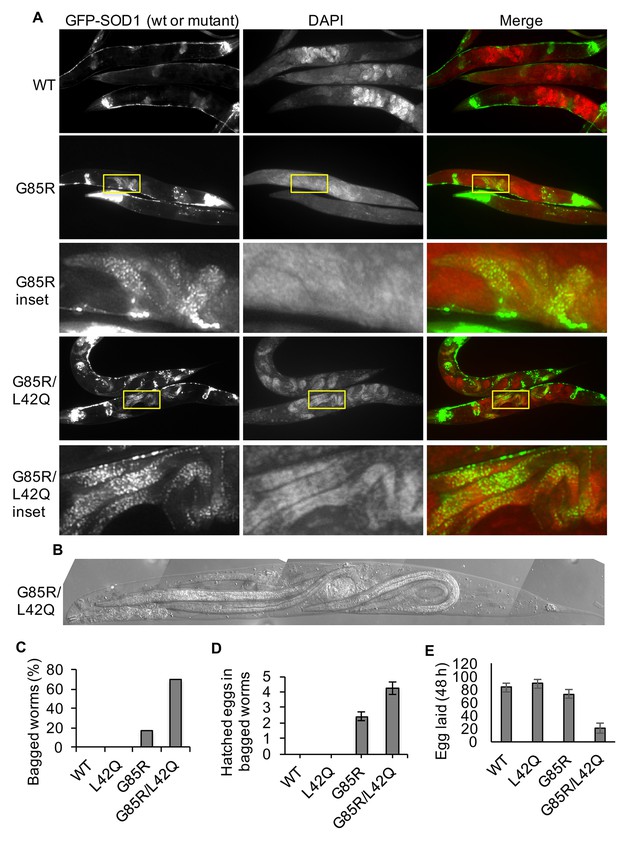
Disruption of the NES by L42Q mutation in SOD1G85R mutant causes severer egg-laying defect in transgenic C. elegans.
(A) Bagging in transgenic worms. Insets show hatched eggs in bagged worms. (B) An example of bagged G85R/L42Q C. elegans. (C) Rates of bagging in transgenic worms. L4 animals were transferred to fresh plates, and bagged worms were counted 48 hr later. n = 46 (WT), n = 45 (L42Q), n = 41 (G85R), n = 47 (G85R/L42Q). (D) Average hatched eggs in bagged worms from (C). n = 7 (G85R), n = 33 (G85R/L42Q). Data represent means±SEM. G85R/L42Q vs G85R: p=0.048 by two-tailed unpaired t-test. (E) Eggs laid in 48 hr. Each L4 animal was transferred to one well of 96-well dish containing S-complete medium with OP50. 48 hr later, larvae (hatched eggs) and unhatched eggs were counted for each well. Data represent means ±SEM. n = 12. G85R vs. G85R/L42Q: p=4.07E-05 by Two-tailed unpaired t-test.
-
Figure 9—source data 1
Data for Figure 9C,D and E.
- https://doi.org/10.7554/eLife.23759.019





