Atomistic simulations indicate the c-subunit ring of the F1Fo ATP synthase is not the mitochondrial permeability transition pore
Figures
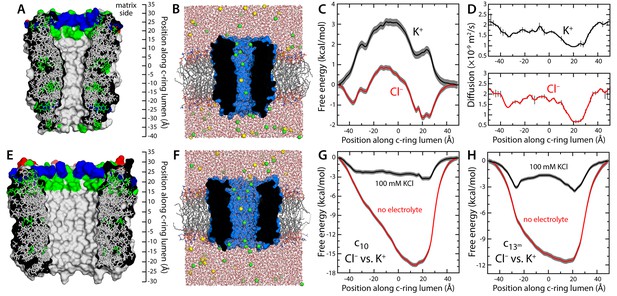
Evaluation of the ion-conducting properties of the c-ring lumen, assuming a hydrated state.
(A) Cross-section of the c10-ring of the mitochondrial ATP synthase from S. cerevisiae. The surface of the protein is colored as follows: Lys and Arg, blue; Asp and Glu, red; other polar residues (and protonated Glu), green; other residues, grey. The ring is oriented such that the interface with the F1 domain, inside the mitochondrial matrix, is up. (B) Molecular simulation system employed to study the properties of the c10-ring, shown again in cross-section (blue). The ring is embedded in a model phospholipid bilayer, in 100 mM KCl. K+ and Cl- ions are shown as yellow and green spheres, respectively. Note the c-ring lumen is hydrated. (C, D) Potential-of-mean-force (G(z), PMF) and diffusion-coefficient profiles for the permeation of either K+ or Cl- across the lumen of the c10-ring (Materials and methods). The lack of binding sites for K+ and the 3 kcal/mol free-energy barrier explain the modest K+ conductance; permeation by Cl-, by contrast, is strongly favored electrostatically. (E, F) Same as (A, B), for a variant of the c13-ring from Bacillus pseudofirmus (Figure 1—figure supplement 2B,C), whose lumen is wider than that of the c10-ring. (G) Free-energy of selectivity for Cl- and against K+ by the c10-ring lumen, examined with and without electrolyte. The selectivity profile, ΔG(z), was calculated by subtracting the individual PMF profiles, G(z), in each case. The marked increase in Cl- selectivity toward the lumen entrance, on the matrix side, confirms the strong electrostatic influence of a ring of arginine residues. (H) Same as (G), for the variant of the c13-ring from B. pseudofirmus. Despite its wider lumen, this ring is also markedly anion selective, unlike the MPTP.
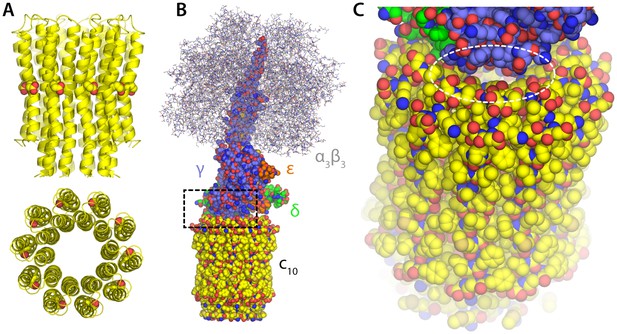
Structure of the F1c10-ring subcomplex of the yeast mitochondrial ATP synthase.
The structure shown corresponds to PDB entry 2WPD (Dautant et al., 2010). Note that the complete F1Fo enzyme includes additional components both inside and outside the membrane, not present in this structure; these missing components sit peripherally and are believed to act as the ‘stator’ against which the moving parts rotate, and also mediate the dimerization of the enzyme. (A) Structure of the c10-ring in the Fo domain, viewed from the plane of the membrane and from the interior of the mitochondrial matrix. Each of the 10 c-subunits consists of two transmembrane helices. The functional proton-binding sites are found on the outermost surface of the ring, roughly halfway across the transmembrane span. (B) The F1 domain sits atop the c-ring and includes the catalytic unit (α3β3) and the so-called central stalk (subunits γ, δ and e), which mechanically couples F1 and Fo. (C) Close-up of the interface between the c-ring and the central stalk, highlighting openings that would plausibly permit the downhill permeation of protons across the c-ring lumen, if this lumen had channel-like characteristics. These openings are considerably w in ATP synthases with larger c-rings (i.e. with greater number of c-subunits).
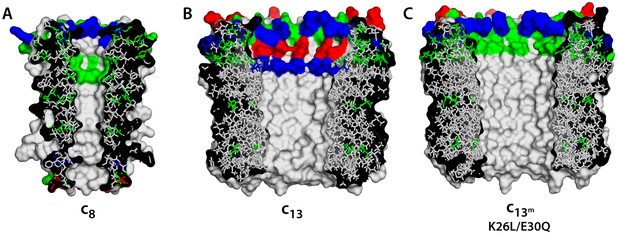
Comparison of c-rings.
(A) Structure of the c8-ring of the ATP synthase from bovine mitochondria (Watt et al., 2010). Side-chains were modeled onto the low-resolution structure. This c8-ring is thought to be conserved across all vertebrates (Watt et al., 2010). (B) Structure of the c13-ring of the ATP synthase from Bacillus pseudofirmus OF4 (Preiss et al., 2010), in the wild-type form. (C) Double-mutant of the c13-ring, made to resemble the lumen of the c8-ring and c10-rings (Figure 1A).
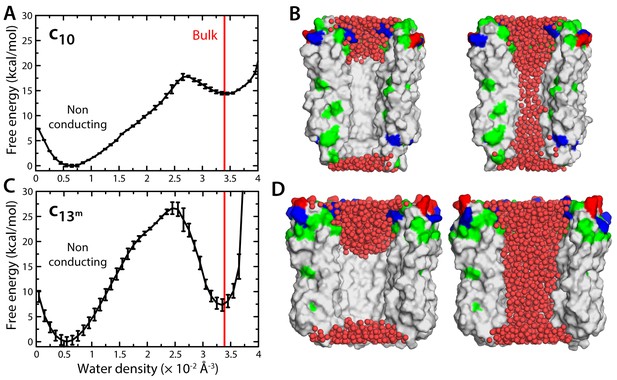
Evaluation of the likelihood of hydration of the c-ring lumen.
(A) Free energy as a function of the water density inside the lumen of the mitochondrial c10-ring, calculated with a variant of the Metadynamics technique (Materials and methods), for a simulation system analogous to that shown in Figure 1B. The density value for bulk water (for the CHARMM36 forcefield) is indicated in red. Error bars reflect the differences between two profiles calculated using different halves of all simulation data. (B) Snapshots of the c-ring lumen in the two metastable minima detected in the free-energy profile shown in panel (A), i.e. a non-conducting, de-wetted state (left), and a water-filled, putatively-conducting state (right), whose properties are characterized in Figure 1. For clarity, several c-subunits in the c-ring are omitted, as are the lipid bilayer and the solvent outside the lumen. Hydrogen atoms in water (red spheres) are also omitted. Note that in the non-conducting state (left), the two regions at the entrance of the hydrophobic portion of the lumen are hydrated; hence, the density value for this state in free-energy profile in panel (A) is not zero. (C, D) Same as (A, B), for the variant of the prokaryotic c13-ring from the B. pseudofirmus ATP synthase.
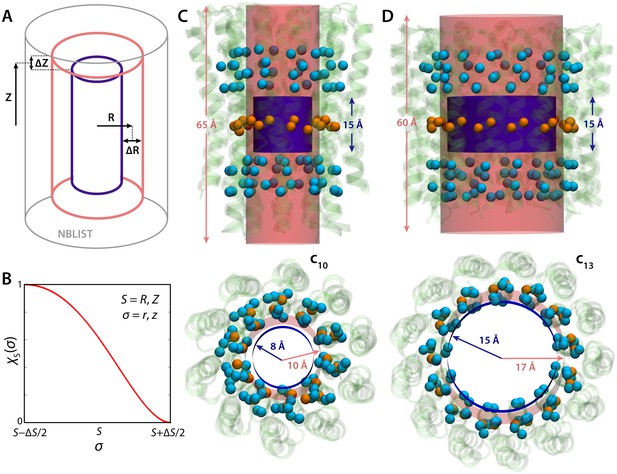
Graphical definition of the density collective-variable with cylindrical geometry.
See Materials and methods. (A) The weight function ξ defines a cylindrical volume with soft boundaries that extend from R-ΔR/2 to R+ΔR/2, and from |Z-ΔZ/2| to |Z+ΔZ/2|. Specifically, the value of ξ inside the blue cylinder is 1, and is 0 outside the red cylinder. In between the two cylinders, ξ switches smoothly from 1 to 0 (panel B). A larger cylinder, shown in grey, is used to define and update a list of particles that neighbor the volume during the simulation. (B) Switching function used to evaluate ξ at the cylinder boundaries. (C, D) Specific definition of the cylindrical volume used in our analysis of the probability of lumen hydration in the c10- and c13-rings. As mentioned, this volume is defined relative to the protein coordinates, and not in absolute Cartesian space; therefore, the volume can change position and orientation during the simulation as the protein tumbles and diffuses within the membrane. To achieve this, three sets of Cα atoms in regions of the protein with minimal structural variability (orange and cyan spheres) are used to define three centers-of-mass. The center of the cylindrical volume is made to coincide and follow the protein center in the middle (orange), while the vector that connects the two distal centers (cyan) defines the cylinder axis.
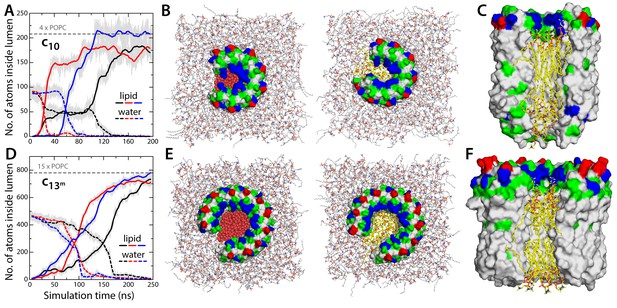
Evidence that lipid molecules block the lumen of the c-ring.
Three independent molecular dynamics simulations were carried out of assembly intermediates of the c10- and c13-rings lacking two of the c-subunits. Initially, the lumen of these c-rings was hydrated at bulk-like density. (A) Time-series of the number of non-hydrogen atoms from either water (dashed lines) or lipid molecules (solid lines) inside the lumen of the mitochondrial c-ring. The volume consider for this count is a cylinder of radius 7 Å and height 32 Å, approximately centered in the middle of the membrane. Running averages are shown for each of the three simulation, colored in black, red and blue, respectively, with the raw data shown in the background in grey. The number of atoms equivalent to 4 POPC lipid molecules is indicated, for reference. (B) Snapshots of the molecular system, at the beginning (left) and at the end (right) of the simulation. Water molecules (red) are progressively displaced by lipid molecules (shown in yellow) entering the lumen laterally. (C) Side-view of the open c-ring at the end of the one of the simulations. The lipid molecules inside the lumen preserve a bilayer arrangement, and adapt to the specific features of the protein surface. (D, E, F) Same as (A, B, C), for the variant of the c13-ring from B. pseudofirmus. The volume considered in panel (D) is a cylinder of radius 14 Å and height 34 Å.





