Insights into electrosensory organ development, physiology and evolution from a lateral line-enriched transcriptome
Figures
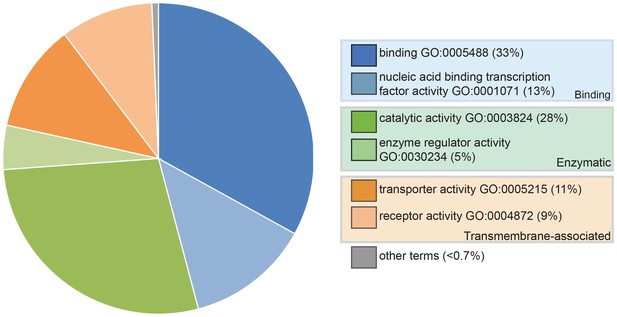
Pie chart showing the results of a PANTHER classification analysis by molecular function of 332 transcripts from the paddlefish lateral line-enriched dataset with associated gene ontology (GO) terms.
The percentage of function hits is indicated in parentheses. Binding activities GO:0005488 (blue) represent ~39% of the function hits (16% nucleic acid binding GO:0003637; 17% protein binding GO:0005515; 6% other binding, including calcium and lipid binding). Catalytic activities GO:0003824 (green) account for ~35% of the function hits (4% enzyme regulator activity GO:0030234; 9% transferase; and 11% each hydrolase and other catalytic activities). Transmembrane-associated activities (orange) represent ~19% of function hits (13% transporter GO:0005215; 6% receptor GO:0004872). The remaining ~7% of function hits are comprised of signal transducer GO:0004871, structural molecule GO:0005198 and antioxidant GO:0016209 activities.
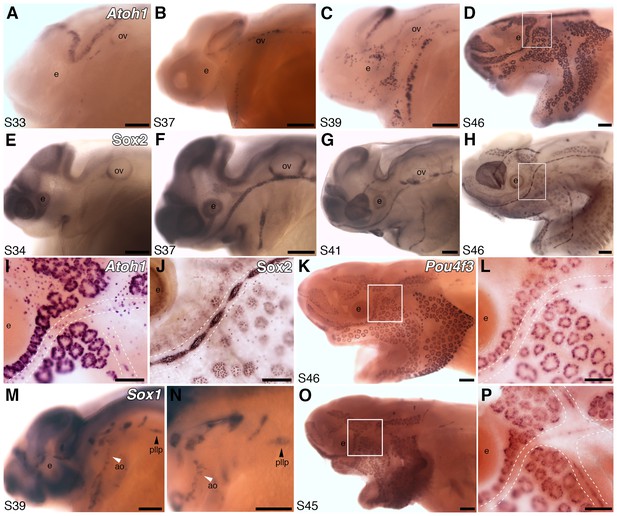
Both ampullary organs and neuromasts express transcription factor genes essential for hair cell development, and selected putative Atoh1 targets.
(A–D) In situ hybridization for paddlefish Atoh1 at stages 33, 37, 39 and 46, respectively. (E–H) Sox2 immunostaining at stages 34, 37, 41 and 46, respectively. (I,J) Higher-power views of stage 46 skin-mounts of Atoh1 (I) and Sox2 (J). Dotted lines indicate approximate boundaries of neuromast containing lateral line canal lines. (K,L) In situ hybridization for Pou4f3 at stage 46 reveals expression in both ampullary organs and neuromasts. (M,N) In situ hybridization for Sox1 at stage 39 shows expression in the posterior lateral line primordium (pllp) and ampullary organs erupting on the operculum. (O,P) At later stages, Sox1 is maintained in ampullary organs and expressed in neuromast canal lines, as shown here at stage 45. Abbreviations: ao, ampullary organ; e, eye; nm, neuromast; ov, otic vesicle, pllp, posterior lateral line primordium. Scale bars: A-H,K,M-O, 200 µm; I,J,L,P, 100 µm.
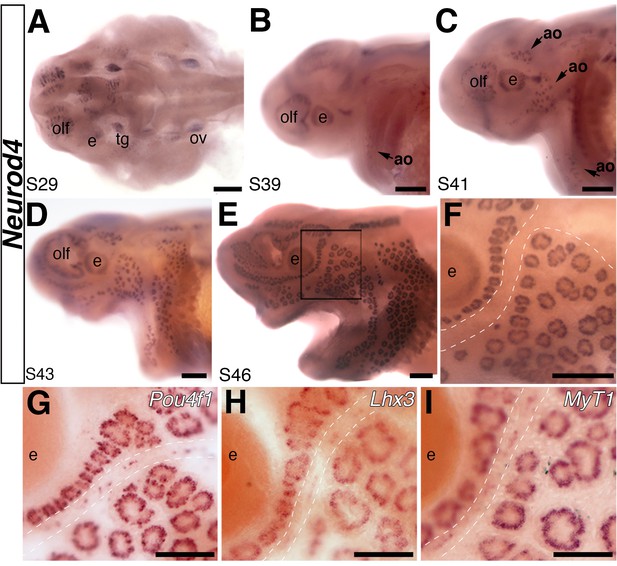
The proneural transcription factor gene Neurod4 is expressed in ampullary organs but not neuromasts.
(A–F) In situ hybridization for Neurod4 at stages 29 (A), 39 (B), 41 (C) 43 (D) and 46 (E). At the earliest stages, transcripts are observed in the olfactory epithelium, eye, trigeminal ganglion. By stage 39 transcripts are observed in the developing ampullary organ fields of the operculum, rapidly expanding to other ampullary organ fields in older embryos. Expression is limited to ampullary organs and is not observed at any stage in neuromasts, as clearly seen at higher power at stage 46 (F). Dotted lines indicate approximate boundaries of neuromast canal lines. (G–I) Potential Neurod4 interactors Pou4f1 (G), Lhx3 (H) and Myt1 (I) are expressed in both ampullary organs and neuromasts. Abbreviations: ao, ampullary organ; e, eye; nm, neuromast; olf, olfactory epithelium; ov, otic vesicle; tg, trigeminal ganglion. Scale bars: A-F, 200 µm; G-I, 100 µm.
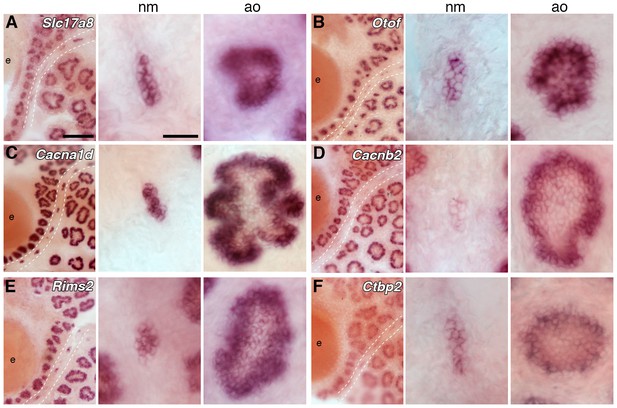
Ampullary organs express genes required for transmission at the hair cell ribbon synapse.
In situ hybridization at stage 46 reveals expression in both ampullary organs and neuromasts of: (A) Slc17a8, encoding the vesicular glutamate transporter 3 (Vglut3); (B) Otof, encoding otoferlin; (C) Cacna1d, encoding the pore-forming alpha subunit of Cav1.3; (D) Cacnb2, encoding an auxiliary beta subunit that is associated with Cav1.3 in hair cells - note that the level of Cacnb2 in neuromasts is weaker than in ampullary organs; (E) Rims2, associated with synaptic ribbons in photoreceptors and hair cells; (F) the Ribeye-specific A domain of Ctbp2, encoding the ribbon-specific protein Ribeye. Scale bars: 100 µm and 20 µm.
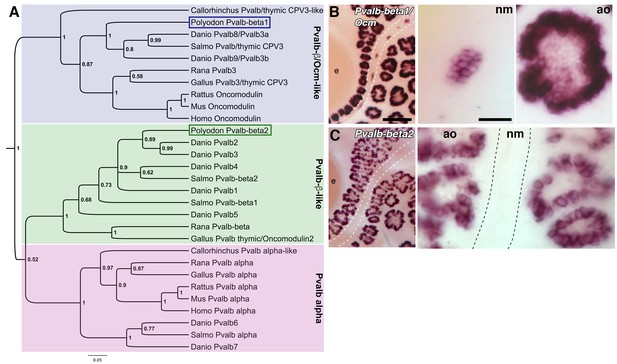
Two beta-parvalbumin genes are differentially expressed in ampullary organs versus neuromasts.
(A) Phylogenetic analysis of selected vertebrate parvalbumin proteins shows three clades: the alpha-parvalbumins, plus two clades containing beta-parvalbumins: the first includes the mammalian beta-parvalbumins, i.e., oncomodulins, and chicken Pvalb3/thymic CPV3. One of the paddlefish lateral line-enriched parvalbumin genes encodes a protein that groups within the beta-parvalbumin clade containing the oncomodulins, so we have named it Pvalbβ1/Ocm. The other falls within the second beta-parvalbumin clade, so we have named it Pvalbβ2. (B) In situ hybridization shows that Pvalbβ1/Ocm is expressed in both ampullary organs and neuromasts, while (C) Pvalbβ2 is restricted to ampullary organs. Scale bars: 100 µm and 20 µm.
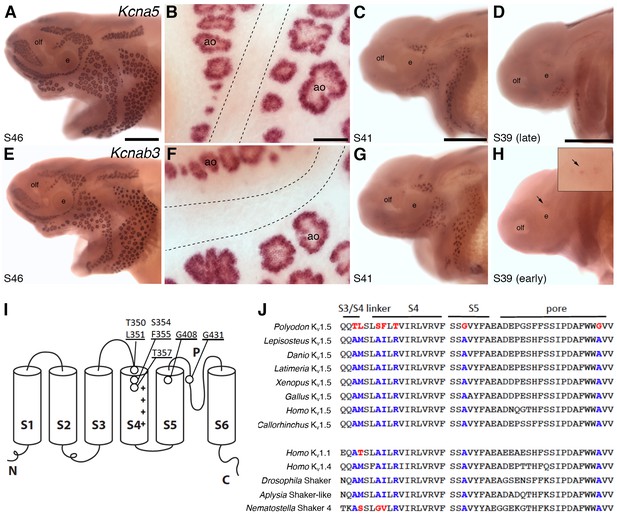
Shaker-related voltage-gated potassium channel subunit genes expressed in ampullary organs but not neuromasts.
(A,B) In situ hybridization at stage 46 for Kcna5, which encodes the pore-forming alpha subunit of the voltage-gated potassium channel Kv1.5, reveals expression only in the developing ampullary organ fields. Dotted lines indicate approximate boundaries of neuromast canal lines. (C,D) Even at the successively earlier stages shown (stage 41 and 39), Kcna5 is still restricted to the developing ampullary organ fields. (E–H) Expression of Kcnab3, encoding the auxiliary beta subunit Kvβ3, is similarly confined to the developing ampullary organ fields. Dotted lines indicate approximate boundaries of neuromast canal lines. The arrow in H, and the higher-power view of this region shown in the inset, indicate the area where the first Kcnab3 expression is noted, at stage 39. Scale bars: A,C-E,G,H, 0.5 mm; B,F, 50 µm. Abbreviations: ao, ampullary organ; e, eye; olf, olfactory pit. (I) Schematic, linear structure of the pore-forming alpha subunit of a Kv channel, with the positions noted of amino acid substitutions in paddlefish Kv1.5. (J) Amino acid sequences across the S3/4 linker region, the voltage-sensing segment S4, plus S5 and the pore, from paddlefish Kv1.5 (top) and other Shaker-related Kv channels for comparison, indicating the deep conservation of some of these amino acid positions across metazoans. The first set of sequences are from Kv1.5 across the jawed vertebrates, including three ray-finned bony fishes: Polyodon spathula (Mississippi paddlefish, a non-teleost chondrostean fish), Lepisosteus oculatus (spotted gar, a non-teleost neopterygian fish), Danio rerio (zebrafish, a teleost neopterygian fish); four lobe-finned fishes/tetrapods: Latimeria chalumnae (coelacanth), Xenopus tropicalis (tropical clawed frog), Gallus gallus (chicken), Homo sapiens (human); and a cartilaginous fish (Callorhinchus milii, a holocephalan). The second set of sequences are from two other human Shaker-related channels (Kv1.1 and Kv1.4), and three invertebrate Shaker orthologs, from Drosophila melanogaster (an insect, i.e., an ecdysozoan), Aplysia californica (a mollusc, i.e., a spiralian) and Nematostella vectensis (a sea anemone, i.e., a cnidarian).
Additional files
-
Supplementary file 1
Excel file listing transcripts that are lateral line-enriched at least 1.85-fold (log2fold 0.89).
- https://doi.org/10.7554/eLife.24197.008





