In vitro FRET analysis of IRE1 and BiP association and dissociation upon endoplasmic reticulum stress
Figures
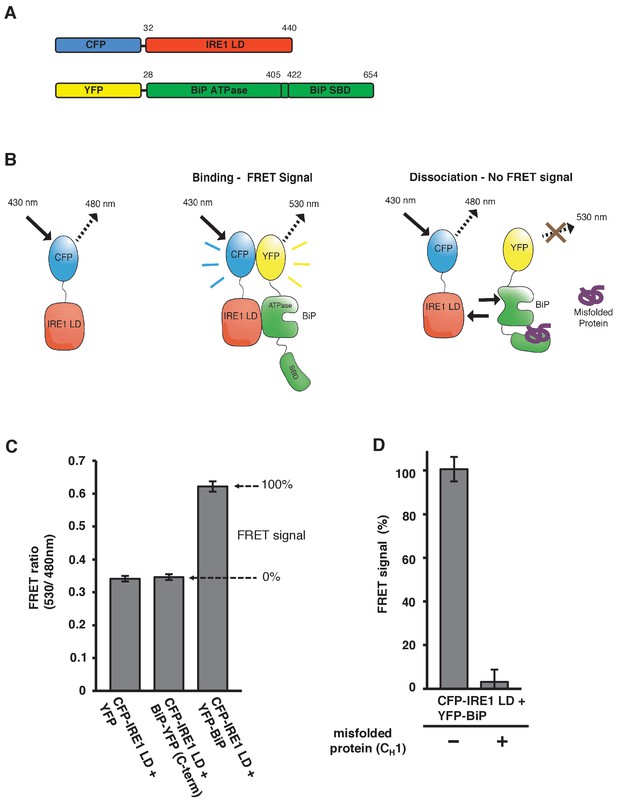
In vitro FRET assay measures noncanonical IRE1 and BiP association and dissociation.
(A) Schematic description of constructs used for the assay, with numbers denoting the amino acid residue of the protein. Both fluorescent proteins, CFP and YFP, are attached N-terminally to the IRE1 LD and BiP proteins, respectively. (B) Schematic representation of the in vitro FRET UPR induction assay, which measures a three-component protein interaction system. CFP-IRE1 LD will be excited at 430 nm with a bandwidth of 10 nm (430–10 nm), and upon excitement it will emit radiation at the longer wavelength of 480–10 nm. When YFP-BiP is added, IRE1 LD should interact with BiP via its ATPase domain, bringing YFP in close proximity to CFP, resulting in FRET between the fluorescent proteins and an emission at 530–10 nm. Dissociation of the complex—with loss of FRET signal—should occur upon addition of misfolded protein, which will bind to BiP substrate-binding domain (SBD) to cause conformational change. The ratio of 530-10 nm/480-10 nm will be used to measure FRET signal output. (C) Bar graph of the FRET ratio (530–10 nm/480–10 nm) upon excitation at 430–10 nm wavelength when CFP-IRE1 LD and YFP-BiP were mixed in equimolar amounts. This was compared to non-binding controls, YFP with CFP-IRE1 LD, and BiP with C-terminally tagged YFP with CFP-IRE1 LD. The FRET ratio was almost doubled upon interaction revealing a clear FRET signal. The negative controls measure a FRET ratio of ~0.34 due to CFP, which contributes a significant fluorescence emission intensity at 530 nm (also referred to as CFP leakage) when excited at 430 nm. This allows for greater spectral overlap with YFP making CFP and YFP excellent FRET pairs, but adds to the background noise. The data are shown as mean ± SD (n = 6). (D) FRET UPR induction assay measurements upon addition of misfolded protein CH1. In this graph, the FRET signal is represented as a percentage, with 0% observation equivalent to non-binding control and 100% being represented by CFP-IRE1 LD and YFP-BiP. The addition of CH1 caused IRE1 LD and BiP dissociation, resulting in the loss of FRET signal (mean ± SD; n = 6).
-
Figure 1—source data 1
- https://doi.org/10.7554/eLife.30257.003
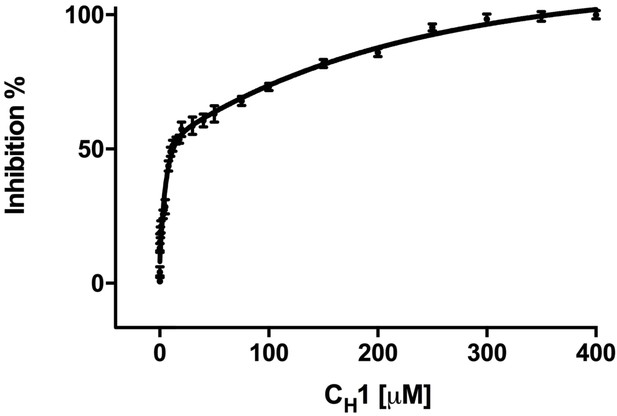
Addition of misfolded protein CH1 inhibits FRET signal in a dose-dependent fashion.
Graph showing the percentage inhibition of FRET signal as a function of the concentration of CH1 (mean ± SD; n = 3). The binding of CH1 to the YFP-BiP caused its dissociation from the CFP-IRE1 LD. At 300 μM CH1, a 10-fold molar excess over both CFP-IRE1 LD and YFP-BiP, the signal was almost completely inhibited. Fitting an exponential (two phase association, r2 = 0.98) curve to data gave an IC50 of 12.3 ± 1.3 μM, the amount of CH1 required to give 50% dissociation, with an inhibition constant Ki = 0.51 ± 0.01 μM. The inhibition constant Ki is the binding constant that relates the interaction between a protein complex with a binding affinity Kd and an inhibiting molecule that is derived from IC50 (Martin et al., 2008).
-
Figure 2—source data 1
- https://doi.org/10.7554/eLife.30257.005
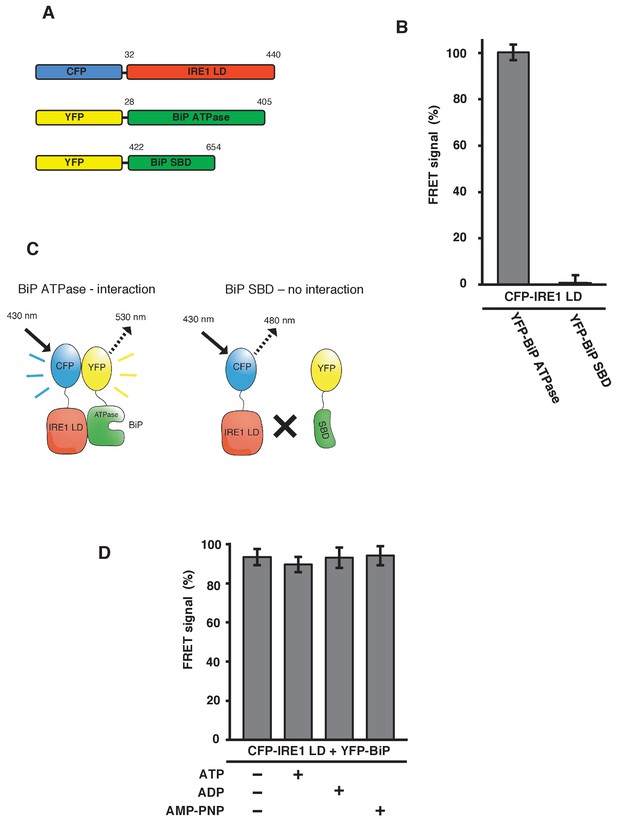
IRE1 LD interacts with the BiP ATPase domain to produce FRET signal, independent of nucleotides.
(A) Diagram detailing the BiP ATPase and SBD constructs used for measuring the interaction with IRE1 LD. (B) IRE1 LD interacted with the BiP ATPase domain to produce 100% FRET signal equivalent to that produced by full-length BiP. No FRET signal observed with the SBD (mean ± SD; n = 6). (C) A schematic illustrating that there was no observable interaction between SBD and IRE1 LD, with no corresponding FRET signal. (D) 5 mM ATP, ADP and AMP-PNP were added to CFP-IRE1 LD and YFP-BiP samples and the FRET signal was analyzed and compared to 100% FRET signal. The addition of nucleotides did not have a significant impact upon the FRET signal and the interaction between IRE1 and BiP.
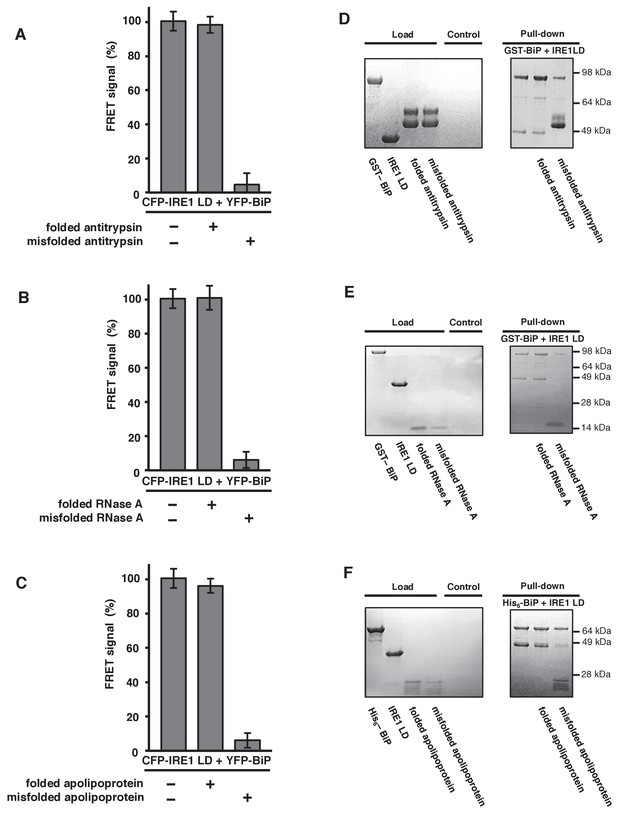
General ER misfolded proteins, – but not their folded state,cause dissociation.
(A–C) Measurements from a FRET UPR induction assay upon addition of folded and misfolded ER proteins. (A) Antitrypsin. (B) RNase A. (C) Apolipoprotein. All data shown are mean ± SD (n = 6). (D–F) Qualitative pull-down assays, stained with coomassie brilliant blue, showed IRE1 LD and BiP dissociation upon binding of a misfolded version of an ER protein. Dissociation did not occur upon the addition of folded proteins. (D) Antitrypsin. (E) RNase A. (F) Apolipoprotein.
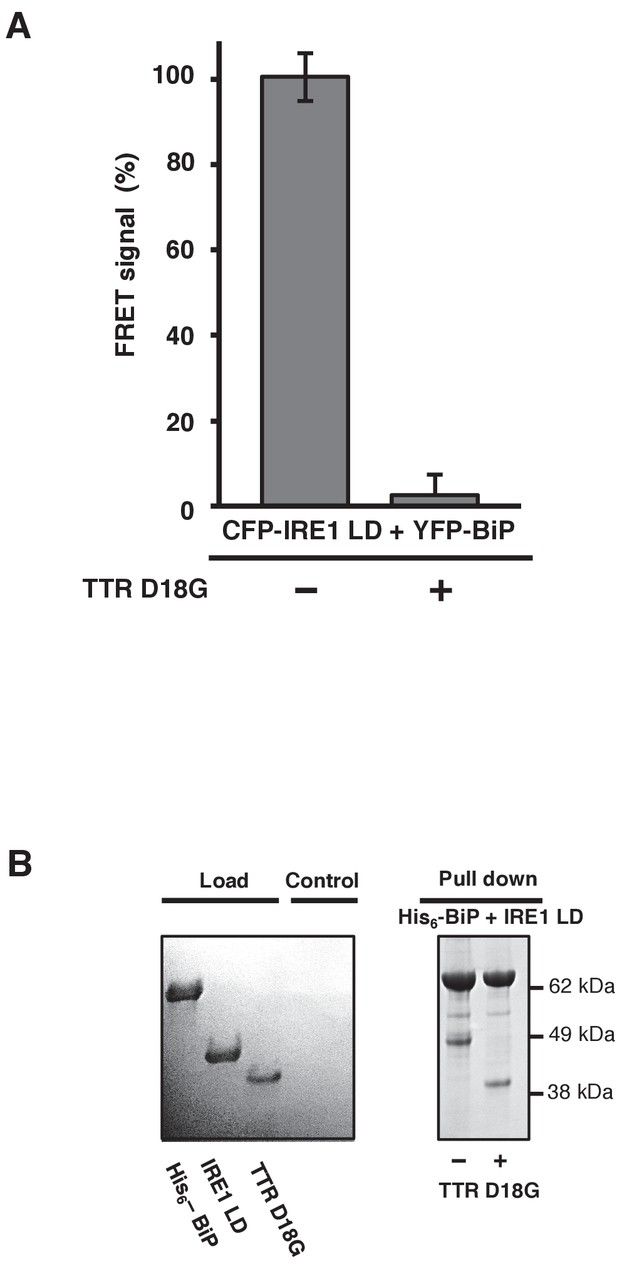
TTRD18G affects IRE1 LD and BiP dissociation.
(A) Transthyretin mutant D18G (TTRD18G) inhibited the FRET signal by causing dissociation of CFP-IRE1 LD and YFP-BiP. (B) A secondary pull-down assay reproduced the results observed with the FRET assay.
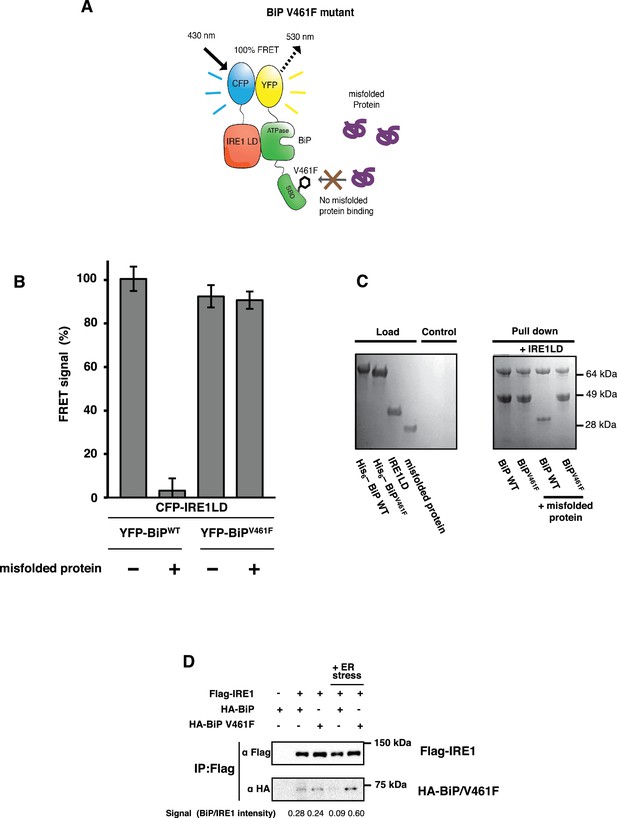
BiP V461F mutation prevents dissociation of the IRE1 LD and BiP upon addition of misfolded protein.
(A) A schematic illustrating the location of the V461F mutation within the SBD of BiP, which is represented by a six-sided black ring that denotes the phenylalanine residue. If the mutation prevents the binding of misfolded protein to BiP, then there will be no allosteric change, and no dissociation of the IRE1–BiP complex, with the FRET signal remaining intact. (B) The FRET signal was significantly reduced upon addition of misfolded protein to BiPWT–IRE1 LD sample. By contrast, there was no loss of FRET signal with BiP V461F. (C) A secondary pull-down assay recapitulates the FRET assay results, with BiPWT fully able to respond to misfolded protein, whereas BiP V461F was unaffected by the presence of misfolded protein. (D) Various combinations of Flag-IRE1, HA-BiP and HA-BiP V461F were co-expressed in HEK293 cells and treated with 5 mM tunicamycin to replicate ER stress. Flag-IRE1 was immunoprecipitated with anti-Flag magnetic resin and then samples were immunoblotted with both Flag and HA antibodies, before and after the addition of ER stress.
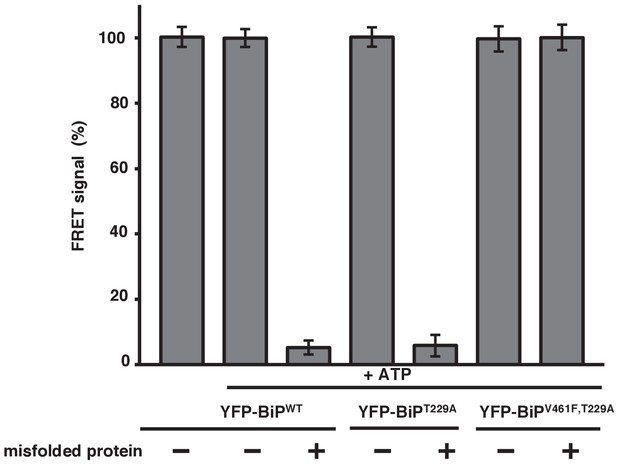
Misfolded proteins are the primary determinant for dissociation of the IRE1 LD and BiP.
In vitro FRET assay showing a comparison between BiPWT and BiPV461F upon the addition of ATP and in the presence of a mutation that renders the BiP ATPase deficient, T229A. Both the T229A mutation and ATP did not affect dissociation. But the presence of the BiP V461F mutation prevented misfolded protein binding with no loss in FRET signal, indicating that misfolded proteins are the primary determinant for dissociation.
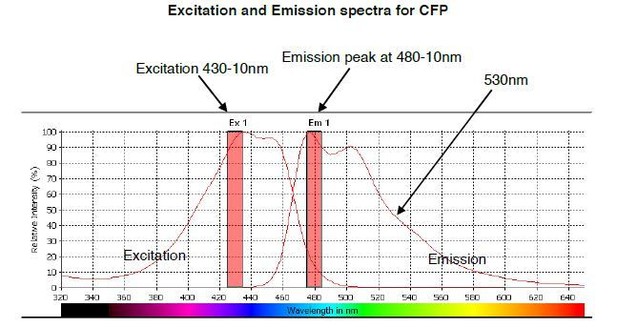
The predicted emission and excitation spectra for CFP when excited at 410 nm with bandwidth 10nm.
The peak emission is measured at 480-10, but there is a significant signal present at 530nm, which is the peak emission point for YFP. The graph was generated by CLARIOstar plate reader BMG labtech.
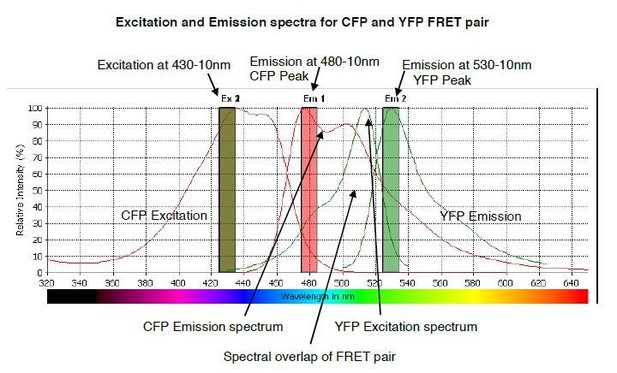
The predicted emission and excitation spectra for CFP and YFP FRET pair when excited at 430-10 nm.
YFP peak emission occurs at 530nm. The graph was generated by CLARIOstar plate reader BMG labtech.
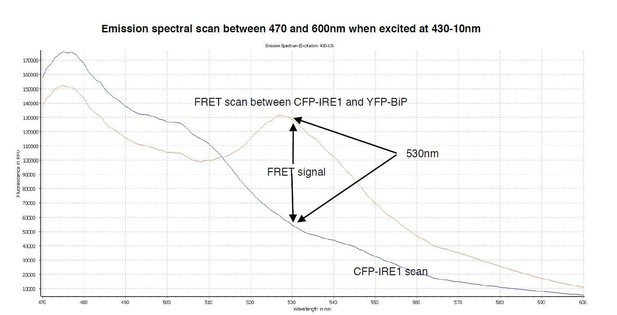
A spectral scan of fluorescence emission intensity between 470nm and 600nm when excited at 430-10nm.
The FRET signal peak at 530nm is clearly visible, also the significant background produced by CFP-IRE1 is observed, producing a background reading of ~0.34 at 530nm- 10/480-10nm wavelength.
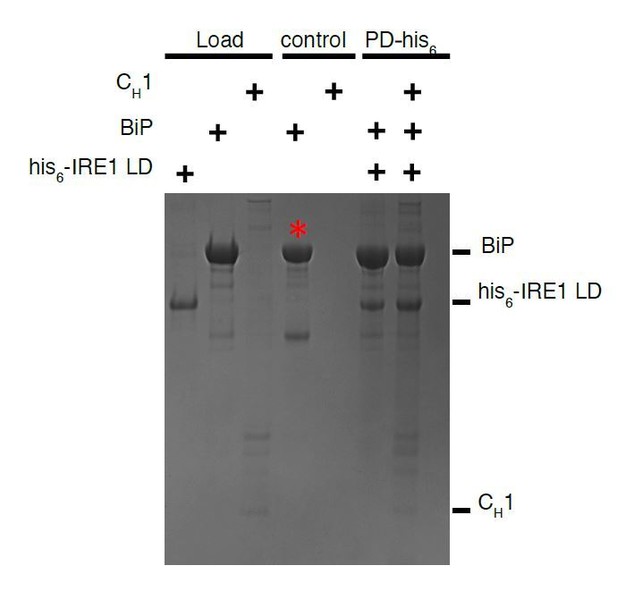
Pull down assay, using his6-tagged IRE1 LD as bait and BiP as prey.
The experiment does not work since un tagged BiP sticks to beads as negative control. The experiment works well when BiP is tagged and using IRE1 LD as prey, since untagged IRE1 LD does not bind beads.
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.30257.011





