Organisms with alternative genetic codes resolve unassigned codons via mistranslation and ribosomal rescue
Figures
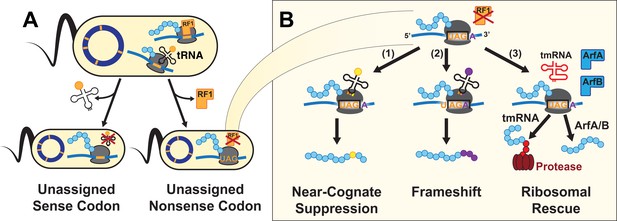
A UAG-ending transcript in the genomically recoded organism (GRO) may produce proteins with multiple differing C-termini.
(A) Unassigned codons arise when either the cognate tRNA or release factor recognizing a codon are removed. (B) Since the GRO lacks Release Factor 1 (RF1), ribosomal stalling at the UAG codons results in three possible fates for the nascent protein (blue): (1) suppression of the codon by a near-cognate or suppressor tRNA (yellow) and continued translation, (2) frameshifting of bases along the mRNA transcript into a new reading frame and continued translation (purple), or (3) ribosomal rescue by the ssrA-encoded tmRNA, ArfA, or ArfB proteins. If ribosomal rescue occurs via tmRNA, the resulting protein is tagged with a peptide sequence (red) for degradation, while rescue via ArfA or ArfB results in release of peptide without C-terminal modification.
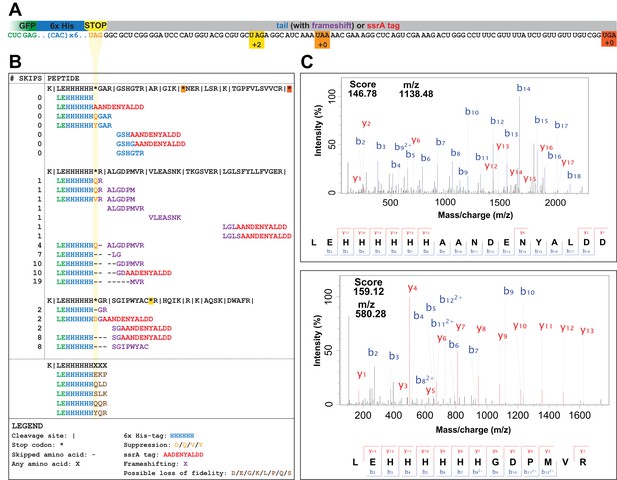
UAG codons in the genomically recoded organism elicit suppression, frameshifting, and tagging for degradation by the tmRNA.
(A) Schematic of the GFP construct with a C-terminal 6x-His tag and a UAG stop codon, showing 102 nucleotides downstream of the UAG codon and the positions of other stop codons in the downstream tail. (B) Peptides identified from the C-terminus of a UAG-ending GFP construct expressed in the GRO (using libraries detailed in Supplementary file 3 and 4). Purified GFP protein was digested with trypsin, processed via MS/MS, and the resulting data were computationally searched using libraries encoding all possible suppressors and all possible subsequent reading frames. Peptides are mapped to the C-terminus of the original GFP construct and grouped by reading frame, with the number of bases skipped listed in the left column. Green text represents GFP, blue text represents the C-terminal 6xHis tag and unframeshifted readthrough, orange text represents the position of a UAG stop codon, purple text represents frameshifted readthrough, and red text represents the tmRNA tag. Black dashes represent ribosomal frameshifts (Figure 2—source datas 1 and 2). (C) MS-MS spectra for two peptides: the C-terminus of GFP with the appended degradation tag (LEHHHHHHAANDENYALDD) and the C-terminus of GFP demonstrating a + 10 base skip in translation (LEHHHHHHGDPMVR). The other spectra validated from UAG-GFP expressing GRO.AA are shown in Supplementary file 2.
-
Figure 2—source data 1
Raw data and analysis of peptides detected in mass spectrometry datasets using a library generated to search for frameshifting, near-cognate suppression, and ribosomal rescue events (Supplementary file 3).
- https://doi.org/10.7554/eLife.34878.005
-
Figure 2—source data 2
Raw data and analysis of peptides detected in mass spectrometry datasets using a library generated to search for loss of translational fidelity (Supplementary file 4).
- https://doi.org/10.7554/eLife.34878.006
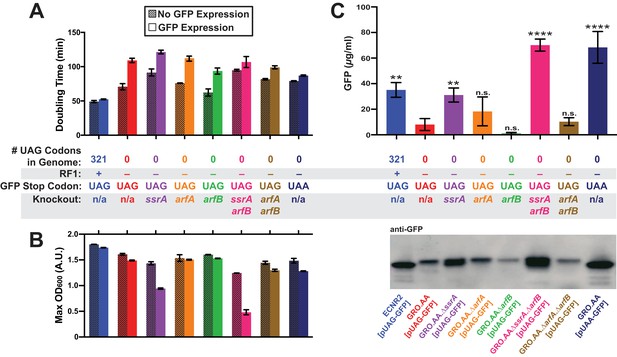
Deletion of both ssrA and arfB restores protein production in the genomically recoded organism.
(A) Comparison of doubling times for WT and GRO strains carrying listed deletions with and without GFP induction. Error bars show standard deviation centered at mean, n = 3; data were analyzed using Source code 1 (Figure 3—source datas 1 and 2). (B) Change in maximum optical density at 600 nm (OD600) due to expression of UAG-GFP or UAA-GFP in wild-type (WT) and GRO strains carrying listed deletions. Error bars show standard deviation centered at mean, n = 3 (Figure 3—source datas 1 and 2). (C) Quantification of GFP abundance per 1 mL of cells at OD600 of 2.5 via western blot from biological replicates of indicated strains (Figure 3—source datas 3–6). Error bars show standard deviation centered at mean, n = 3 (Figure 3—source datas 3–5). See Figure 3—figure supplement 1 for linear calibration curves used to quantify GFP abundance for each replicate experiment. Image of representative western blot is below the graph. p-values are calculated in relation to the GRO containing the UAG-ending GFP (GRO – UAG) and are as follows: * is p≤0.05, ** is p≤0.01, *** is p≤0.001, and **** is p≤0.0001.
-
Figure 3—source data 1
Growth curve data from 96-well plate assay analyzed using Source code 1 (one of three plate replicates), used for data represented in Figure 3A and B.
- https://doi.org/10.7554/eLife.34878.011
-
Figure 3—source data 2
Analysis of doubling times and maximum OD600’s of indicated strains.
File contains doubling times and maximum OD600’s for three separate experiments conducted on different plate reader machines. Each experiment tested each sample in biological triplicate. Only the biological triplicate data from Plate 3 is represented in Figure 3A and B.
- https://doi.org/10.7554/eLife.34878.012
-
Figure 3—source data 3
Anti-GFP western blot image used for quantification of GFP yields; replicate 1.
- https://doi.org/10.7554/eLife.34878.013
-
Figure 3—source data 4
Anti-GFP western blot image used for quantification of GFP yields; replicate 2.
- https://doi.org/10.7554/eLife.34878.014
-
Figure 3—source data 5
Anti-GFP western blot image used for quantification of GFP yields; replicate 3.
- https://doi.org/10.7554/eLife.34878.015
-
Figure 3—source data 6
Analysis of western blot data represented in Figure 3C.
- https://doi.org/10.7554/eLife.34878.016
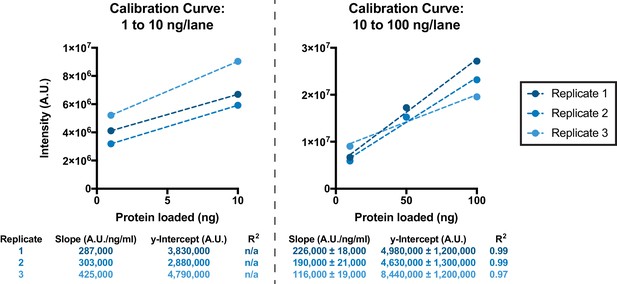
Calibration curves used for quantification of GFP yields, as represented in Figure 3C, using GFP samples of known concentration.
Replicate 1 corresponds to the western blot shown in Figure 3—source data 3; Replicate 2 corresponds to the western blot shown in Figure 3—source data 4; Replicate 3 corresponds to the western blot shown in Figure 3—source data 5.
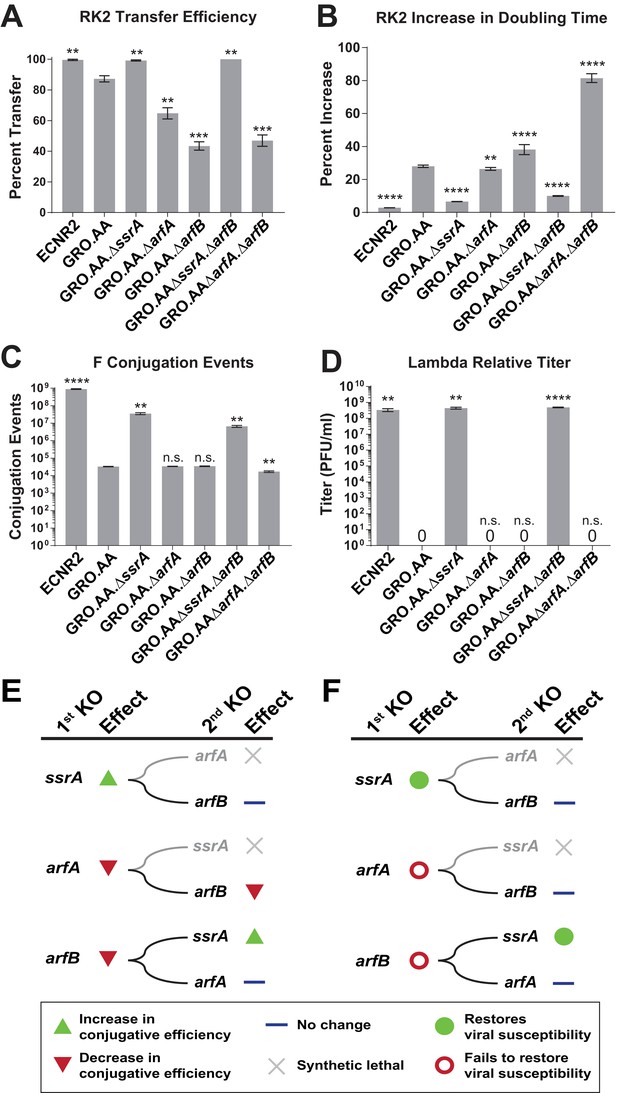
Deleting ssrA restores propagation of both viruses and conjugative plasmids in the genomically recoded organism.
(A) Percent transfer of conjugative plasmid RK2 from a wild-type donor into wild-type (WT), GRO, or GRO with designated deletions (KO) as recipients (Figure 4—source data 1). Data are obtained from technical triplicates generated from a single biological sample. (B) Percent increase in doubling time for strains carrying plasmid RK2 compared to strains lacking RK2 (Figure 4—source datas 2 and 3). (C) Number of conjugation events for conjugative plasmid F from wild-type, GRO, or GRO with designated gene deletions as donors to a wild-type recipient (Figure 4—source data 4). Data are obtained from technical triplicates generated from a single biological sample. (D) Relative titer on wild-type, GRO, and GRO with designated deletions of phage λ (Figure 4—source data 5). Error bars show standard deviation centered at mean, n = 3. p-values are calculated in relation to the GRO condition and are as follows: * is p≤0.05, ** is p≤0.01, *** is p≤0.001, and **** is p≤0.0001. (E) Effects of sequential deletions of ribosomal rescue mechanisms on conjugative plasmid transfer efficiency. (F) Effects of sequential deletions of ribosomal rescue mechanisms on viral susceptibility.
-
Figure 4—source data 1
Analysis of RK2 plasmid conjugation data represented in Figure 4A.
Note: These data represent technical triplicates generated from the same biological sample.
- https://doi.org/10.7554/eLife.34878.018
-
Figure 4—source data 2
Growth curve data from 96-well plate assay analyzed using Source code 1, used for data represented in Figure 4B.
- https://doi.org/10.7554/eLife.34878.019
-
Figure 4—source data 3
Analysis of doubling times represented in Figure 4B.
- https://doi.org/10.7554/eLife.34878.020
-
Figure 4—source data 4
Analysis of F plasmid conjugation data represented in Figure 4C.
Note: These data represent technical triplicates generated from the same biological sample.
- https://doi.org/10.7554/eLife.34878.021
-
Figure 4—source data 5
Analysis of lambda phage infection data represented in Figure 4D.
- https://doi.org/10.7554/eLife.34878.022
Tables
Strains used in this study.
https://doi.org/10.7554/eLife.34878.007| Strain Abbreviation* | Ancestor (source)† | Genotype | # UAG Codons‡ | RF1 Status§ | Ribosomal rescue gene deletion | ssrA tag Status# | Investigated in |
|---|---|---|---|---|---|---|---|
| GRO.DD.prfA+ | GRO.AA (this study) | ΔmutS:zeo.Δ(ybhB-bioAB):[λcI857.Δ(cro-ea59):tetR-bla] | 0 | +RF1 | n/a | DD | GFP expression for mass spectrometry (Figure 2) |
| GRO.DD | GRO.AA (this study) | ΔmutS:zeo.Δ(ybhB-bioAB):[λcI857.Δ(cro-ea59):tetR-bla], ΔprfA, ΔtolC | 0 | ∆RF1 | n/a | DD | GFP expression for mass spectrometry (Figure 2) |
| ECNR2.AA | E. coli MG1655 (Wang et al., 2009) | MG1655 ΔmutS:zeo.Δ(ybhB-bioAB):[λcI857.Δ(cro-ea59):tetR-bla] | 321 | +RF1 | n/a | AA | Fitness, conjugation, and viral infection (Figures 3 and 4) |
| GRO.AA | ECNR2.AA (Lajoie et al., 2013b) | ΔmutS:zeo.Δ(ybhB-bioAB):[λcI857.Δ(cro-ea59):tetR-bla], ΔprfA, ΔtolC | 0 | ∆RF1 | n/a | AA | Fitness, conjugation, and viral infection (Figures 3 and 4) |
| GRO.AA.∆ssrA | GRO.AA (this study) | ΔmutS:zeo.Δ(ybhB-bioAB):[λcI857.Δ(cro-ea59):tetR-bla], ΔprfA, ΔtolC | 0 | ∆RF1 | ∆ssrA | AA | Fitness, conjugation, and viral infection (Figures 3 and 4) |
| GRO.AA.∆arfA | GRO.AA (this study) | ΔmutS:zeo.Δ(ybhB-bioAB):[λcI857.Δ(cro-ea59):tetR-bla], ΔprfA, ΔtolC | 0 | ∆RF1 | ∆arfA | AA | Fitness, conjugation, and viral infection (Figures 3 and 4) |
| GRO.AA.∆arfB | GRO.AA (this study) | ΔmutS:zeo.Δ(ybhB-bioAB):[λcI857.Δ(cro-ea59):tetR-bla], ΔprfA, ΔtolC | 0 | ∆RF1 | ∆arfB | AA | Fitness, conjugation, and viral infection (Figures 3 and 4) |
| GRO.AA.∆ssrA.∆arfB | GRO.AA (this study) | ΔmutS:zeo.Δ(ybhB-bioAB):[λcI857.Δ(cro-ea59):tetR-bla], ΔprfA, ΔtolC | 0 | ∆RF1 | ∆ssrA, ∆arfB | AA | Fitness, conjugation, and viral infection (Figures 3 and 4) |
| GRO.AA.∆arfA. ∆arfB | GRO.AA (this study) | ΔmutS:zeo.Δ(ybhB-bioAB):[λcI857.Δ(cro-ea59):tetR-bla], ΔprfA, ΔtolC | 0 | ∆RF1 | ∆arfA, ∆arfB | AA | Fitness, conjugation, and viral infection (Figures 3 and 4) |
-
*All strains derived from ECNR2, as described in Wang et al. (2009).
†See Key Resources Table for additional information on strains and sources. The GenBank accession number for E. coli MG1655 is U00096, and the GenBank accession number for GRO.AA is CP006698.
-
‡ Out of a total of 321 in the original ECNR2 strain.
§RF1 terminates translation at UAG and UAA. Deletion of RF1 eliminates recognition of UAG during translation; translational termination continues through RF2, which recognizes UAA and UGA.
-
#The ssrA gene encodes the tmRNA, which appends the ssrA degradation tag to stalled ribosomes. The wild-type sequence is AANDENYALAA; mutation of the C-terminus to AANDENYALDD slows degradation of peptides to enable detection by mass spectrometry.
Components of peptide library constructed to search and analyze tandem mass spectrometry data.
The LEHHHHHHXXX library was separate from the library that contained the entries of the first three rows of the table (see Supplementary file 3 and 4).
| Library component | Example peptides (from Figure 2A) | Enables detection of… | Complete peptide list |
|---|---|---|---|
| Any one of 20 canonical amino acids inserted at the UAG codon | LEHHHHHHQGAR | Near-cognate suppression | Supplementary file 3 |
| Any length of C-tail following UAG codon to the next non-UAG stop codon or to 38 amino acids downstream of the UAG codon, whichever came first | ALGDPMVR | Readthrough, frameshifting, and rescue by ArfA or ArfB | Supplementary file 3 |
| AANDENYALDD degradation tag | LEHHHHHHGDAANDENYALDD | Rescue by tmRNA-SmpB | Supplementary file 3 |
| All peptides of form LEHHHHHHXXX, where X is any amino acid | LEHHHHHHQLD | Loss of translational fidelity | Supplementary file 4 |
Genetic reagents, bacterial strains, antibodies, and software used in this study.
https://doi.org/10.7554/eLife.34878.023| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information | Isaacs Lab Reference # | Full genotype of strains | # UAG Codons | RF1 status | Ribosomal rescue gene knockout | ssrA tag status |
|---|---|---|---|---|---|---|---|---|---|---|
| Gene (Escherichia coli) | pUAG-GFP | this paper | eGFP-6xHis -UAG; Plasmid NJM88; Strain NJM1242 | eGFP protein with a C-terminal 6-His tag for protein purification, terminating translation in a UAG codon. | Plasmid NJM88; Strain NJM1242 | N/A | N/A | N/A | N/A | N/A |
| Gene (E. coli) | pUAA-GFP | this paper | eGFP-6xHis -UAA; Plasmid NJM89; Strain NJM1249 | eGFP protein with a C-terminal 6-His tag for protein purification, terminating translation in a UAA codon. | Plasmid NJM89; Strain NJM1249 | N/A | N/A | N/A | N/A | N/A |
| Genetic reagent (E. coli) | RK24 | 10.1126/science .1205822; 10.1016/j.cels .2016.06.009 | pRK24; Strain NJM699 | Conjugative RK2 plasmid (10.1006/ jmbi.1994.1404), but lacks functional AmpR gene. | Strain NJM699 | N/A | N/A | N/A | N/A | N/A |
| Genetic reagent (E. coli) | F | Yale University Coli Genetic Stock Center (CGSC), Strain #4401 | pF; Strain EMG2; Strain CGSC#4401; Strain NJM426; Strain NJM473 | Conjugative F plasmid, as described by PMID: 4568763. Obtained from the Yale CGSC. | Strain NJM426; Strain NJM473 | N/A | N/A | N/A | N/A | N/A |
| Genetic reagent (E. coli) | pZE21_ UAG-GFP | this paper | pZEtR-eGFP -cHis-TAG- v02; Plasmid NJM88; Strain NJM1242 | pZE21 plasmid with pLtetO promoter driving inducible expression of eGFP with a C-terminal 6-His tag and terminating in UAG codon. Inducible with anhydro-tetracycline. | Plasmid NJM88; Strain NJM1242 | N/A | N/A | N/A | N/A | N/A |
| Genetic reagent (E. coli) | pZE21_ UAA-GFP | this paper | pZEtR-eGFP -cHis-TAA-v02 ; Plasmid NJM89; Strain NJM1249 | pZE21 plasmid with pLtetO promoter driving inducible expression of eGFP with a C-terminal 6-His tag and terminating in UAA codon. Inducible with anhydro-tetracy cline. | Plasmid NJM89; Strain NJM1249 | N/A | N/A | N/A | N/A | N/A |
| Genetic reagent (Enteroba cteria phage λ) | λ.CI857 | Coli Genetic Stock Center (CGSC), Yale University (contact John Wertz directly) | λ.CI857; λ phage; Phage NJM102 | Phage λ with temperature- sensitive CI repressor gene; when incubated at 37° C, phage becomes obligate lytic | Phage NJM102 | N/A | N/A | N/A | N/A | N/A |
| Cell line (E. coli) | GRO.DD | this paper | C31GIB. tmRNA-DD; Strain #987 | MG1655-derived strain with all 321 UAG codons mutated to UAA, deletion of RF1, and tmRNA tag C-terminal amino acids mutated from AA to DD. Retains lambda red cassette for recombineering. Investigated in Figure 2. | Strain #987 | ΔmutS:zeo. Δ(ybhB- bioAB) :[λcI857. Δ(cro-ea59) :tetR-bla]. ΔprfA.ΔtolC .tmRNADD | 0 | +RF1 | n/a | DD |
| Cell line (E. coli) | GRO. DD.prfA+ | this paper | C31GIB. prfA+.tmRNA -DD; Strain #996 | MG1655-derived strain with all 321 UAG codons mutated to UAA, retains RF1, and tmRNA tag C-terminal amino acids mutated from AA to DD. Retains lambda red cassette for recombineering. Investigated in Figure 2. | Strain #996 | ΔmutS:zeo. Δ(ybhB- bioAB) :[λcI857. Δ(cro-ea59): tetR-bla]. ΔtolC.tm RNADD | 0 | ∆RF1 | n/a | DD |
| Cell line (E. coli) | ECNR2 | 10.1016/j.cels .2016.06.009 | ECNR2.Δmut S:zeocin.Δ λRed; Strain #795 | MG1655-derived strain that contains 321 UAG codons and retains RF1. Investigated in Figures 3 and 4. | Strain #795 | ΔmutS:zeo | 321 | +RF1 | n/a | AA |
| Cell line (E. coli) | GRO.AA | 10.1016/j.cels .2016.06.009 | C31.final. ΔmutS: zeocin.ΔprfA .ΔλRed; Strain #796 | MG1655-derived strain with all 321 UAG codons mutated to UAA, deletion of RF1. Investigated in Figures 3 and 4. | Strain #796 | ΔmutS: zeo.ΔprfA (GenBank ID: CP006698) | 0 | ∆RF1 | n/a | AA |
| Cell line (E. coli) | GRO. AA.∆arfB | this paper | C31GIB.arfB: tolCorf. ΔλRed; Strain #1230 | MG1655-derived strain with all 321 UAG codons mutated to UAA, deletion of RF1, and deletion of arfB. Investigated in Figures 3 and 4. | Strain #1230 | ΔmutS: zeo.ΔprfA .arfB:tolC | 0 | ∆RF1 | ∆ssrA | AA |
| Cell line (E. coli) | GRO. AA.∆ssrA | this paper | C31GIB.ssrA :tolC.ΔλRed; Strain #1231 | MG1655-derived strain with all 321 UAG codons mutated to UAA, deletion of RF1, and deletion of ssrA. Investigated in Figures 3 and 4. | Strain #1231 | ΔmutS: zeo.ΔprfA. ssrA:tolC | 0 | ∆RF1 | ∆arfA | AA |
| Cell line (E. coli) | GRO. AA.∆arfA | this paper | C31GIB.arfA :tolC.ΔλRed ; Strain #1232 | MG1655-derived strain with all 321 UAG codons mutated to UAA, deletion of RF1, and deletion of arfA. Investigated in Figures 3 and 4. | Strain #1232 | ΔmutS: zeo.ΔprfA. arfA:tolC | 0 | ∆RF1 | ∆arfB | AA |
| Cell line (E. coli) | GRO.AA .∆ssrA.∆arfB | this paper | C31GIB.ΔarfB .ssrA:tolC.Δ λRed; Strain #1233 | MG1655-derived strain with all 321 UAG codons mutated to UAA, deletion of RF1, and deletion of ssrA and arfB. Investigated in Figures 3 and 4. | Strain #1233 | ΔmutS: zeo.ΔprfA .ΔarfB.ssrA:tolC | 0 | ∆RF1 | ∆ssrA. ∆arfB | AA |
| Cell line (E. coli) | GRO.AA .∆arfA. ∆arfB | this paper | C31GIB.Δarf B.arfA:tolC. ΔλRed; Strain #1234 | MG1655-derived strain with all 321 UAG codons mutated to UAA, deletion of RF1, and deletion of arfA and arfB. Investigated in Figures 3 and 4. | Strain #1234 | ΔmutS: zeo.ΔprfA .ΔarfB.arfA :tolC | 0 | ∆RF1 | ∆arfA. ∆arfB | AA |
| Antibody | mouse anti-GFP antibody | other | Invitrogen (Ref#: 332600, Lot#: 1513862A) | Invitrogen (Ref#: 332600, Lot#: 1513862A); (5.5 μL antibody in 3 mL Milk + TBST) | N/A | N/A | N/A | N/A | N/A | N/A |
| Antibody | goat anti-mouse antibody | other | AbCam (Ref#: ab7023, Lot#: GR157827-1) | AbCam (Ref#: ab7023, Lot#: GR157827-1); (2.2 μL antibody in 10 mL Milk + TBST) | N/A | N/A | N/A | N/A | N/A | N/A |
| Recombinant DNA reagent | ssrA:tolC | this paper; for use, see tolC positive /negative selection in 10.1038/nprot .2014.081 | dsDNA NJM111 | The E. coli native tolC gene used to delete ssrA gene via recombineering (10.1038/nprot. 2008.227). | dsDNA NJM111 | N/A | N/A | N/A | N/A | N/A |
| Recombinant DNA reagent | arfA:tolC | this paper; for use, see tolC positive /negative selection in 10.1038/nprot .2014.081 | dsDNA NJM112 | The E. coli native tolC gene used to delete arfA gene via recombineering (10.1038/nprot. 2008.227). | dsDNA NJM112 | N/A | N/A | N/A | N/A | N/A |
| Recombinant DNA reagent | arfB:tolC | this paper; for use, see tolC positive /negative selection in 10.1038/nprot .2014.081 | dsDNA NJM113 | The E. coli native tolC gene used to delete arfB gene via recombineering (10.1038/nprot. 2008.227). | dsDNA NJM113 | N/A | N/A | N/A | N/A | N/A |
| Software, algorithm | Doubling time algorithm | 10.1126/ science.1241459 | Growth_ Analyze_ GK.m | Doubling time used in 10.1126 /science.1241459, written by Gleb Kuznetsov in the lab of Dr. George Church. | N/A | N/A | N/A | N/A | N/A | N/A |
| Software, algorithm | MaxQuant v1.5.1.2 | other | N/A | Commercial software for mass spectrometry analysis. | N/A | N/A | N/A | N/A | N/A | N/A |
| Software, algorithm | Graphpad Prism 7 | other | N/A | Commercial software for statistical analysis and graphing, provided through Yale University. | N/A | N/A | N/A | N/A | N/A | N/A |
Additional files
-
Source code 1
MATLAB script used to analyze growth curve data from 96-well plate assays (Figure 3A,B and 4B).
- https://doi.org/10.7554/eLife.34878.024
-
Supplementary file 1
Gene nucleotide sequences, processed mass spectrometry data, and numerical values used to generate Figures 2–4.
- https://doi.org/10.7554/eLife.34878.025
-
Supplementary file 2
Spectra for all 47 manually verified peptides detected through mass spectrometry from GRO.AA expressing the UAG-GFP plasmid.
- https://doi.org/10.7554/eLife.34878.026
-
Supplementary file 3
Library of peptides generated for the detection of frameshifting, near-cognate suppression, and ribosomal rescue events from mass spectrometry data.
- https://doi.org/10.7554/eLife.34878.027
-
Supplementary file 4
Library of peptides generated for the detection of loss of translational fidelity from mass spectrometry data.
- https://doi.org/10.7554/eLife.34878.028
-
Transparent reporting form
- https://doi.org/10.7554/eLife.34878.029





