Comment on ‘YcgC represents a new protein deacetylase family in prokaryotes’
Figures
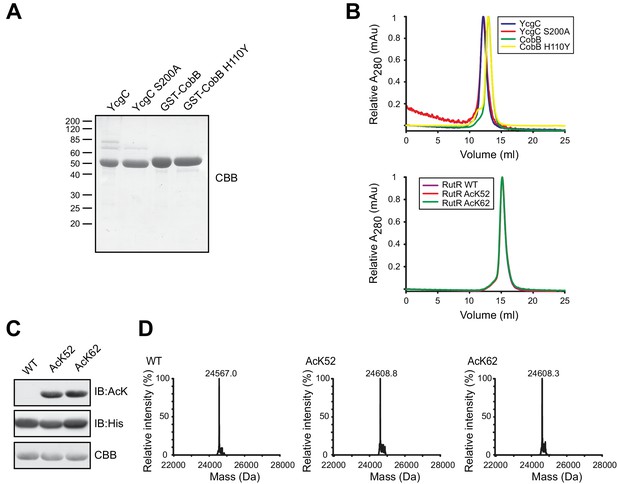
Preparation of YcgC, CobB and non-acetylated/acetylated RutR.
(A) SDS-PAGE analysis of proteins used in this study. All proteins were expressed and purified as GST-fusion proteins. In terms of YcgC and YcgC S200A the GST-tag was removed by TEV protease during the purification steps. Staining of the gel was done by coomassie brilliant blue (CBB). For molecular masses see figure legend for Figure 2A. (B) Analytical size exclusion chromatography on a S200 10/300 GL column shows that YcgC WT and YcgC S200A as well as CobB and the corresponding catalytically inactive variant CobB H110Y display an almost identical elution profile. Moreover, RutR proteins show a nearly identical elution profile in analytical SEC runs indicating that RutR acetylation at K52 and K62 does not interfere with protein folding or its oligomeric state. (C) RutR-His6 AcK52 and AcK62 are quantitatively acetylated. Shown are SDS-PAGE and immunoblot analyses of all RutR-His6 proteins used in this study. Staining for AcK using an anti-acetyl-L-lysine antibody revealed a strong signal for RutR AcK52 and AcK62, whereas no signal was obtained for RutR WT. As loading control anti-His6 staining was performed. (D) ESI-MS data show the quantitative and homogenous incorporation of acetyl-L-lysine into RutR. Shown is the deconvoluted spectrum on the true mass scale after software transformation yielding one single peak and the corresponding molecular mass as indicated. Expected mass non-acetylated RutR: 24567.5 Da; acetylated RutR: 24609.5 Da.
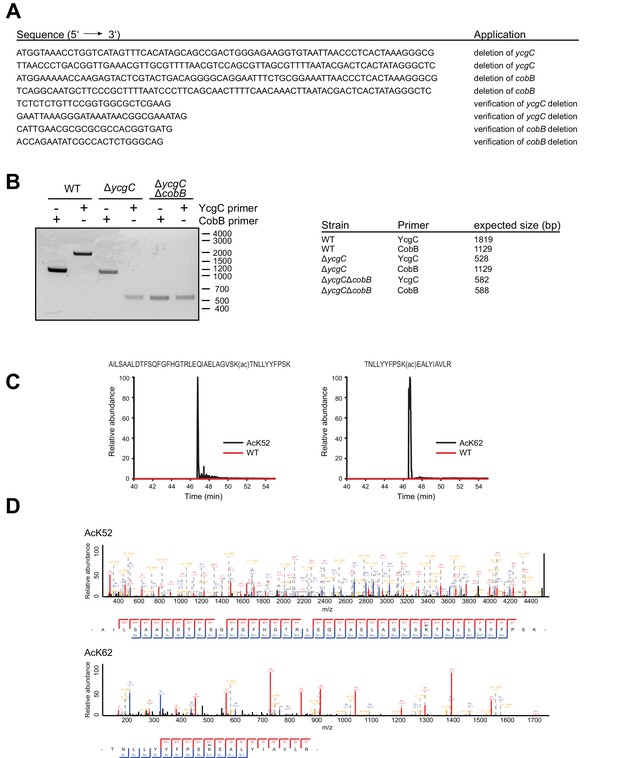
Construction of an E.coli BL21 (DE3) ΔycgCΔcobB double knockout strain and tryptic digest and subsequent LC-MS analysis of RutR AcK52 and RutR AcK62.
(A) Oligonucleotides used to generate an E. coli BL21 (DE3) strain with genomic deletion of ycgC and cobB genes. (B) Validation of gene deletions in E. coli BL21 (DE3) ΔycgCΔcobB by PCR. Shown is the agarose gel of PCR products obtained from different E. coli strains. Application of oligonucleotides that bind 200 bp up- and downstream of the ycgC or cobB gene confirm successful deletions. (C) Full MS scan chromatograms of listed RutR peptides. Peptide intensities validate the successful incorporation of acetyllysine at positions K52 and K62, respectively. Corresponding acetylated peptides were only rarely found in RutR WT. (D) MS/MS spectra of acetylated RutR peptides as stated.
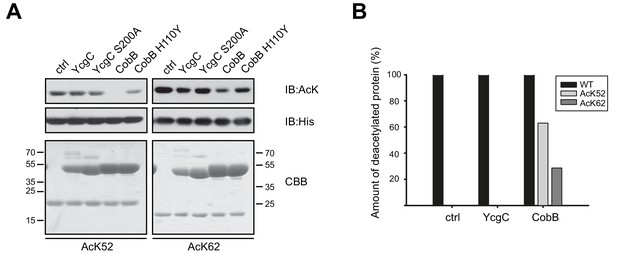
YcgC does not show any deacetylase activity towards RutR AcK52 or RutR AcK62 and it does not stimulate RutR (auto-) proteolytic cleavage.
(A) RutR AcK52 and RutR AcK62 were treated with a two-fold molar excess of YcgC, YcgC S200A, CobB or CobB H110Y in the presence of 5 mM NAD+. As visible from the immunoblotting using an anti-AcK AB CobB was able to completely deacetylate RutR AcK52 while RutR AcK62 is only faintly deacetylated under the conditions used. YcgC, YcgC S200A and the catalytically dead CobB H110Y did neither deacetylate RutR AcK52 nor RutR AcK62 suggesting that YcgC is no deacetylase for RutR. We used the GST-fusion proteins of CobB and CobB H110Y here to get a better separation of CobB and RutR proteins. Probing of the His6-tag using an anti-His6 antibody was done to show equal loading with RutR proteins. SDS-PAGE analysis and CBB staining show that there is only one band of the same size for RutR for all the conditions tested suggesting that YcgC did also not stimulate (auto-) proteolytic cleavage of RutR. The control lane (ctrl) shows the respective RutR protein, AcK52 or AcK62, without addition of CobB or YcgC. Molecular weights of proteins used (all masses calculated without N-terminal methionine): RutR, 24567.5 Da; acetylated RutR, 24609.5 Da; GST-CobB, 53271.09 Da; GST-CobB H110Y, 53297.12 Da.; YcgC, 51649.80 Da; YcgC S200A, 51633.80 Da. We used the GST-fusion proteins for CobB to obtain a better separation from RutR as CobB (MW: 26314.86 Da) and CobB H110Y (26340.90 Da) without GST-tag have similar molecular weights compared to RutR. (B) Quantification of ESI-MS spectra of YcgC and CobB reaction products shown as immunoblots in (A). As a support for the data obtained by immunoblotting, neither YcgC nor YcgC S200A nor catalytically inactive CobB H110Y did alter the molecular mass of RutR WT, AcK52 and AcK62. However, active CobB led to more than 60% deacetylation of RutR AcK52 while RutR AcK62 was only marginally deacetylated (app. 30% deacetylated). Again, these data confirm that YcgC does neither deacetylate or proteolytically cleave RutR nor does it stimulate autoproteolytic cleavage of RutR.

Identification and quantification of acetylated and deacetylated RutR by ESI-MS.
RutR incubated with or without YcgC and CobB as stated in the experimental section was tested for acetylation by ESI-MS molecular weight determination. Shown are deconvoluted spectra on the true mass scale after software transformation. Obtained peaks are labelled with calculated masses each. The calculated masses correspond to the expected masses for RutR WT (24567.5 Da) and RutR AcK52/AcK62 (24609.5 Da).
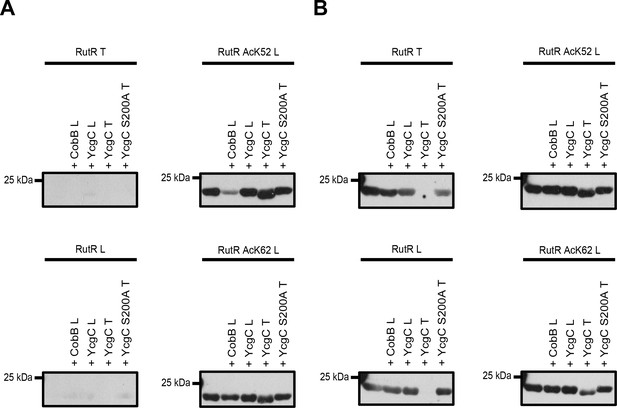
Comparison of YcgC and RutR proteins as well as antibody detection from Tu et al.
(T) and from our lab (L). (A) RutR from Tu et al. (RutR T) and our non-acetylated RutR (RutR L) were treated with YcgC prepared by our lab (L), YcgC and YcgC S200A from Tu et al. (T). RutR T and non-acetylated RutR L are not detected by the antibody ab21623 from abcam (left panels). Site-specifically lysine-acetylated RutR AcK52 and AcK62 prepared by us (RutR AcK52 L and RutR AcK62 L) are detected by the antibody ab21623. Treatment with CobB reduces acetylation level for RutR AcK52 L and AcK62 L as expected (right panels). Treatment of RutR AcK52 L and RutR AcK62 L with YcgC T, but not with YcgC S200A T and YcgC L, results in reduction of molecular size, while acetylation level is not decreased. (B) Same as in A but staining was done with anti acetyl-lysine antibody CST9441 from Cell Signaling Technology. CST9441 antibody detects site-specifically acetylated and non-acetylated RutR (or RutR that contains highly sub-stoichiometric, trace amounts of lysine acetylation). Acetylation signal is completely removed for RutR T upon treatment with YcgC T but neither with YcgC S200A nor with YcgC L. His6-staining shows that YcgC T treatment also removes N-terminal His6-tag suggesting proteolytic cleavage from N-terminus (Figure 3—figure supplement 1B). RutR AcK52 L and AcK62 L also show reduction in molecular size upon treatment with YcgC T (right panels). However, acetylation signal using CST9441 antibody of the degradation band is not removed, showing that deacetylation at neither RutR AcK52 L nor RutR AcK62 L takes place. CobB treatment of no RutR protein results in visible reduction in acetylation signal using CST9441 antibody, although it is shown that CobB deacetylates AcK52 and AcK62 from RutR.
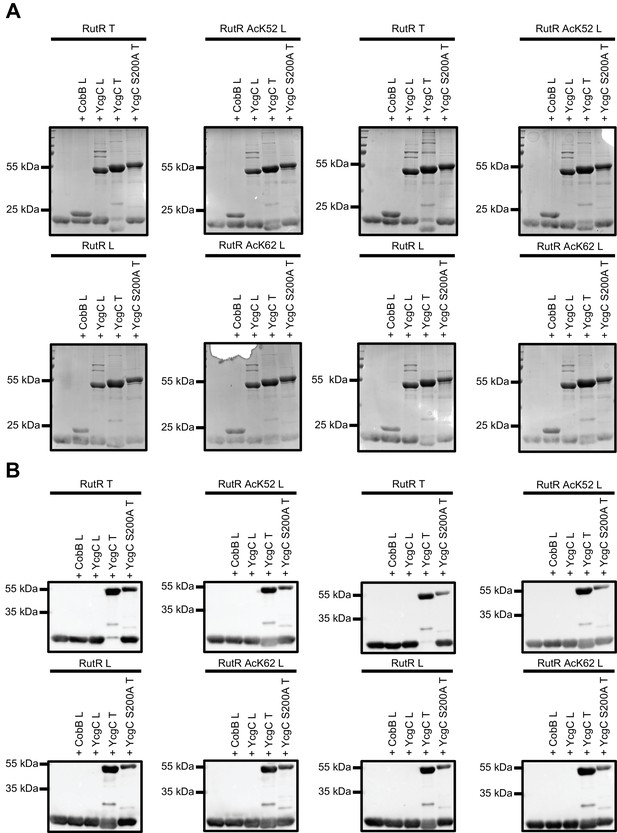
Loading controls of the deacetylation assays performed under conditions described by Tu et al.
The reactions shown in Figure 3 were stained with Coomassie Brilliant Blue (A) and anti-His6-antibody (B). Notably, treatment of N-terminally His6-tagged RutRT by YcgC T almost completely removes anti-His6-signal, while all C-terminally tagged RutR L proteins (non-acetylated, AcK52 and AcK62) show downshift to lower molecular size but still generate a signal using anti-His6-antibody, suggesting N-terminal proteolytic cleavage of RutR by YcgC T (or by a proteolytic activity that resides in YcgC preparation from Tu et al.).
Additional files
-
Supplementary file 1
The dataset contains all proteins identified in preparations of recombinantly expressed YcgC and YcgC S200A from Tu et al. and of YcgC prepared by us.
- https://doi.org/10.7554/eLife.37798.008
-
Supplementary file 2
Proteases identified in the YcgC sample of Tu et al., which are neither present in the catalytically inactive YcgC S200A preparation from Tu et al. nor in the YcgC preparation from Kremer and Kuhlmann et al.
- https://doi.org/10.7554/eLife.37798.009
-
Supplementary file 3
The dataset contains both all proteins identified in preparation of recombinantly expressed RutR from Tu et al. as well as acetylated and non-acetylated peptides from RutR from Tu et al.
We observed an overall sequence coverage of almost 93%. The only sequences not identified were 8-TTGKRSRAVSAK-19 and 100-LK-101, most likely because trypsin cleavage generates small peptides not detected by LC-MS/MS. As a result, this shows that there is no acetylation occurring at those lysines.
- https://doi.org/10.7554/eLife.37798.010
-
Supplementary file 4
The YcgC amino acid sequence was analysed by the threading programme I-iTASSER.
Usage of the structural alignment programme TM-align to match the first I-TASSER model to all structures in the PDB. The following table shows the top ten proteins that show the closest structural similarity to the predicted I-TASSER model. Due to the structural similarity these proteins often have similar function as the target.
- https://doi.org/10.7554/eLife.37798.011
-
Supplementary file 5
Protein sequences of recombinantly expressed RutR and YcgC proteins from Tu et al.
- https://doi.org/10.7554/eLife.37798.012





