Brain Networks: Modeling breathing rhythms
Orchestral music is beautiful, rich and complex, yet analyzing the contribution of any one instrument can be difficult. In many ways, neuroscientists face similar challenges when they try to understand how the networks in the brain work. These circuits are made of many types of neurons, each of which has different properties. Moreover, the properties of a neuron can change because of its interactions with other cells in the network.
Researchers have started to unravel this complexity by building computational models of both neurons and networks of neurons, and by focusing on pairs of interacting properties. This is similar to how one might study how the left and right hand of a violinist work together to produce melody and rhythm, and then use this knowledge to better understand other stringed instruments and their roles in the orchestra.
Within the ‘rhythmogenic’ networks of the brain, different types of neurons work together to create the body rhythms that are essential for life. For example, a complex network generates breathing rhythms, and it is often used to understand rhythmogenic circuits in general. The balance between excitatory and inhibitory connections between neurons is critical to shape network activity (Ramirez and Baertsch, 2018). Equally important, but less well understood, are the interactions between intrinsic properties that are built into individual neurons.
In particular, two intrinsic properties are thought to play a role in controlling breathing rhythms and network activity in general. The first is a persistent sodium current (INaP), which is slowly activated and inactivated by changes in the voltage across the neuronal membrane. The second does not depend on voltage: rather, the calcium-activated non-selective cation current (ICAN) is triggered when the level of calcium ions inside the cell increases. The source of these calcium ions is unknown, but there is experimental evidence that they could be provided by mechanisms at the synapses between neurons (Del Negro et al., 2010; Del Negro et al., 2011). Both INaP and ICAN allow some neurons to generate rhythms on their own, without being stimulated by their neighbors. Yet, in the actual network, these properties also enhance the signals transmitted by excitatory synapses (Ramirez et al., 2004). Because INaP and ICAN interact with synaptic properties, as well as with each other, it is difficult to isolate their relative contributions.
Pharmacological and genetic manipulations have shed light on how INaP and ICAN work in the respiratory network (see, for example, Koizumi et al., 2018; Picardo et al., 2019), but there is still no consensus on how they contribute to rhythmogenesis (Feldman and Del Negro, 2006; Ramirez et al., 2004). Now, in eLife, Jeffrey Smith of the National Institute Neurological Disorders Stroke (NINDS) and colleagues – including Ryan Phillips as first author – report results from a computational modeling approach that re-examines how INaP and ICAN control breathing (Phillips et al., 2019).
The researchers, who are based at NINDS, the University of New Hampshire and Georgia State University, established a simplified model of the breathing network, which only takes into account excitatory synaptic interactions. Then, they tuned the contributions of three variables, INaP, ICAN, and the source of intracellular calcium ions, and explored how this affected the frequency and amplitude of breathing. First, they examined how changes in the source of calcium ions influenced the contribution of ICAN to the network. If calcium came from within neurons, ICAN controlled the frequency, but not the strength, of breathing. On the other hand, if calcium depended on synaptic activity, ICAN acted as a synaptic amplifier to control the strength of the rhythm, but it had little effect on its frequency. This scenario best matched experimental data and prior conclusions (Del Negro et al., 2011; Koizumi et al., 2018), prompting Phillips et al. to conclude that ICAN is activated by calcium ions that are primarily of synaptic origin.
The group then went on to demonstrate that ICAN does not establish the rhythm in their computational model, but that it increases the strength of breathing by recruiting more neurons to participate in the network. Indeed, when ICAN was eliminated from the model network, a weak breathing rhythm remained, which was generated by a small number of neurons with high levels of INaP activity (Figure 1). Eliminating this ‘rhythmogenic kernel’ from the model stopped the rhythm altogether, in agreement with some (Peña et al., 2004), but not all (Pace et al., 2007) experimental data.
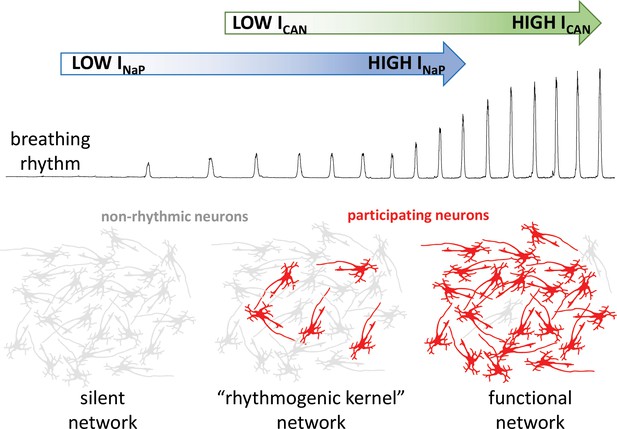
How the intrinsic properties of neurons contribute to the breathing rhythm.
Phillips et al. established a model of the brain network that controls breathing, and used it to deduce how two built-in properties of neurons, the INaP sodium current (blue) and the ICAN current (green), control the breathing rhythm. Without INaP, the network is silent, with the neurons exhibiting non-rhythmic activity (grey). However, a small number of neurons with high levels of INaP activity can produce a weak rhythm, even in the absence of ICAN. In turn, the model suggests that ICAN is activated by calcium ions coming from synapses; the current would then amplify excitatory interactions between neurons. This amplification leads to additional neurons participating in the rhythm (red), producing a robust functional network.
Unlike the model built by Phillips et al., the actual network that controls breathing is not exclusively excitatory, but is subjected to important inhibitory and neuromodulatory control. Further, alternative mechanisms of rhythmogenesis that do not depend on INaP have also been proposed (Del Negro et al., 2010; Rubin et al., 2009). Could different rhythmogenic mechanisms therefore contribute to breathing, depending on the demands of the network? In particular, could the contributions of INaP and ICAN change based on the neuromodulatory state of the network? Addressing such questions will require further back-and-forth between experiments and increasingly complex models.
Overall, the results of Phillips et al. allow us to understand how intrinsic neuronal properties independently control the strength and frequency of the breathing rhythm. Their model is also a useful framework in which to explore how changes in the way INaP and ICAN interact can dynamically impact rhythmogenic properties. For example, it could shed light on the way how the network reconfigures when the body lacks oxygen. Ultimately, describing the duet between INaP and ICAN in the respiratory network may help dissect how rhythmic activity is controlled in other regions of the brain (Penn et al., 2016; Riquelme et al., 2018).
References
-
Dendritic calcium activity precedes inspiratory bursts in preBötzinger complex neuronsJournal of Neuroscience 31:1017–1022.https://doi.org/10.1523/JNEUROSCI.4731-10.2011
-
Looking for inspiration: new perspectives on respiratory rhythmNature Reviews Neuroscience 7:232–241.https://doi.org/10.1038/nrn1871
-
Role of persistent sodium current in mouse preBötzinger complex neurons and respiratory rhythm generationThe Journal of Physiology 580:485–496.https://doi.org/10.1113/jphysiol.2006.124602
-
Pacemaker neurons and neuronal networks: an integrative viewCurrent Opinion in Neurobiology 14:665–674.https://doi.org/10.1016/j.conb.2004.10.011
-
Subcellular localization and activity of TRPM4 in medial prefrontal cortex layer 2/3Frontiers in Cellular Neuroscience 12:12.https://doi.org/10.3389/fncel.2018.00012
Article and author information
Author details
Publication history
- Version of Record published: March 25, 2019 (version 1)
Copyright
© 2019, Ramirez and Baertsch
This article is distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use and redistribution provided that the original author and source are credited.
Metrics
-
- 1,202
- views
-
- 179
- downloads
-
- 1
- citations
Views, downloads and citations are aggregated across all versions of this paper published by eLife.
Download links
Downloads (link to download the article as PDF)
Open citations (links to open the citations from this article in various online reference manager services)
Cite this article (links to download the citations from this article in formats compatible with various reference manager tools)
Further reading
-
- Computational and Systems Biology
Transcriptomic profiling became a standard approach to quantify a cell state, which led to accumulation of huge amount of public gene expression datasets. However, both reuse of these datasets or analysis of newly generated ones requires significant technical expertise. Here we present Phantasus - a user-friendly web-application for interactive gene expression analysis which provides a streamlined access to more than 96000 public gene expression datasets, as well as allows analysis of user-uploaded datasets. Phantasus integrates an intuitive and highly interactive JavaScript-based heatmap interface with an ability to run sophisticated R-based analysis methods. Overall Phantasus allows users to go all the way from loading, normalizing and filtering data to doing differential gene expression and downstream analysis. Phantasus can be accessed on-line at https://alserglab.wustl.edu/phantasus or can be installed locally from Bioconductor (https://bioconductor.org/packages/phantasus). Phantasus source code is available at https://github.com/ctlab/phantasus under MIT license.
-
- Computational and Systems Biology
- Evolutionary Biology
A comprehensive census of McrBC systems, among the most common forms of prokaryotic Type IV restriction systems, followed by phylogenetic analysis, reveals their enormous abundance in diverse prokaryotes and a plethora of genomic associations. We focus on a previously uncharacterized branch, which we denote coiled-coil nuclease tandems (CoCoNuTs) for their salient features: the presence of extensive coiled-coil structures and tandem nucleases. The CoCoNuTs alone show extraordinary variety, with three distinct types and multiple subtypes. All CoCoNuTs contain domains predicted to interact with translation system components, such as OB-folds resembling the SmpB protein that binds bacterial transfer-messenger RNA (tmRNA), YTH-like domains that might recognize methylated tmRNA, tRNA, or rRNA, and RNA-binding Hsp70 chaperone homologs, along with RNases, such as HEPN domains, all suggesting that the CoCoNuTs target RNA. Many CoCoNuTs might additionally target DNA, via McrC nuclease homologs. Additional restriction systems, such as Type I RM, BREX, and Druantia Type III, are frequently encoded in the same predicted superoperons. In many of these superoperons, CoCoNuTs are likely regulated by cyclic nucleotides, possibly, RNA fragments with cyclic termini, that bind associated CARF (CRISPR-Associated Rossmann Fold) domains. We hypothesize that the CoCoNuTs, together with the ancillary restriction factors, employ an echeloned defense strategy analogous to that of Type III CRISPR-Cas systems, in which an immune response eliminating virus DNA and/or RNA is launched first, but then, if it fails, an abortive infection response leading to PCD/dormancy via host RNA cleavage takes over.






