Oxygen Sensors: When is a target not a target?
Oxygen is essential for life – just count how many times you need to breathe while reading this article – and is used by virtually every cell in the human body. Most cells are able to sense a diminished oxygen supply (hypoxia) and respond by making changes to cellular metabolism, blood vessel formation and oxygen delivery. For example, physiological hypoxia, such as that encountered in anemia or at high altitudes, induces the production of a hormone called EPO, which causes the body to make more red blood cells to improve oxygen delivery.
Cells must have highly responsive oxygen sensors to regulate these processes, but to date researchers have found only one family of enzymes – the 2-oxoglutarate dioxygenase enzyme family – that is capable of sensing physiological levels of oxygen. These enzymes use molecular oxygen (O2), iron ions and 2-oxogluterate (a molecule with the chemical formula C5H6O5) to catalyze the transfer of oxygen onto amino acid or DNA substrates (Islam et al., 2018). They act as both hydroxylases, catalyzing the addition of a hydroxyl group (OH) to substrates, and dioxygenases, using O2 as a cosubstrate. Researchers have identified four enzymes from this family that act as oxygen sensors to regulate three transcription factors called HIF1α, HIF2α and HIF3α, which in turn regulate how cells express genes in response to hypoxia. Three of these enzymes are prolyl hydroxylase (PHD) enzymes, and the fourth is called factor inhibiting HIF (Epstein et al., 2001; Ivan et al., 2001; Hewitson et al., 2002; Lando et al., 2002).
The PHD enzymes use molecular oxygen to catalyze the hydroxylation of two proline amino acids in the HIFα proteins. When oxygen levels are normal, the HIFα proteins are hydroxylated, which causes them to be degraded by the cell (Figure 1). However, when oxygen levels decrease, leading to hypoxia, the HIFα proteins are not hydroxylated, so they are not degraded as rapidly. This allows them to migrate to the nucleus and activate the genes responsible for adapting to hypoxia (Kaelin and Ratcliffe, 2008).
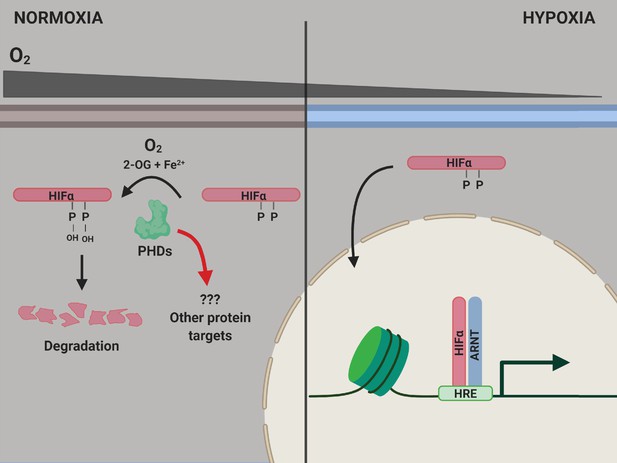
PHD enzymes, HIFα protein and the hypoxia response.
When oxygen levels are normal (normoxia, left), a PHD enzyme (green) can use molecular oxygen (O2), iron ions (Fe2+) and 2-oxogluterate (2-OG) to hydroxylate (ie, add an OH group to) two proline amino acids (P) on a HIFα protein. Hydroxylation destabilizes the HIFα protein, causing it to be degraded by the cell, and the genes involved in the hypoxic response of the cell are not expressed. When oxygen levels are low (hypoxia, right), the PHD enzyme is not able to hydroxylate the HIFα protein, so this protein can migrate into the nucleus and bind to a protein called ARNT. Together, they interact with hypoxia response elements (HREs) in the genome to activate the transcription of hypoxia response genes. ARNT: aryl hydrocarbon receptor nuclear translocator or hypoxic inducible factor-β (HIFβ); HIF: hypoxic inducible transcription factor; PHD: prolyl hydroxylase.
Since the discovery of the PHD enzymes and factor inhibiting HIF, it has been unclear whether these enzymes could hydroxylate targets other than the HIFα proteins. If PHD enzymes hydroxylate other targets it would suggest that additional non-HIF pathways might be involved in the hypoxia response. Previous research efforts have identified many other potential targets for the PHD enzymes, including some with links to physiological responses to hypoxia (Strowitzki et al., 2019). However, many of these studies did not demonstrate that the PHD enzymes were directly catalyzing the hydroxylation of these proteins, raising doubts as to whether these proteins are bona fide targets for the PHD enzymes. Now, in eLife, Matthew Cockman (Francis Crick Institute), Peter Ratcliffe (University of Oxford) and co-workers – including Kerstin Lippl and Ya-Min Tian (both in Oxford) as joint first authors with Cockman, and other researchers from the Crick, Oxford and the University of Oulu – report on a fascinating study that seeks to clarify the situation (Cockman et al., 2019).
Cockman et al. undertook a rigorous array of in vitro biochemical and mass spectrometry experiments using purified enzymes and substrates. While they confirmed that the PHD enzymes robustly catalyze the hydroxylation of proline residues in HIFα proteins, they found no evidence for the hydroxylation of any of the other targets in vitro. Overall, they studied more than 20 different candidate target proteins and 40 potential modification sites. The substrates used in the experiments were short synthetic peptides and full-length recombinant proteins.
These results suggest that the HIFα proteins are the only primary targets of the oxygen-sensing PHD enzymes. If this is the case, then PHD enzymes have a more focused role in hypoxic signaling than previously thought. This is important for predicting the consequences of manipulating PHD activity for therapeutic purposes. However, while these well-controlled, designed and executed biochemical experiments show that targets other than HIFα proteins cannot be efficiently hydroxylated in vitro, they do not preclude the modification of these targets in vivo. This is because living cells might contain additional cofactors that the PHD enzymes need to hydroxylate non-HIFα targets. Furthermore, modulating the activity of the PHD enzymes affects HIF-independent processes, indirectly pointing to potential non-HIF targets. (Strowitzki et al., 2019).
If in vivo experiments confirm that the HIFα proteins are the only primary targets of the PHD enzymes, as Cockman et al. suggest, this would make these enzymes central to of one of the most specialized sensing and control systems in the cell.
References
-
Hypoxia-inducible factor (HIF) asparagine hydroxylase is identical to factor inhibiting HIF (FIH) and is related to the cupin structural familyJournal of Biological Chemistry 277:26351–26355.https://doi.org/10.1074/jbc.C200273200
-
2-Oxoglutarate-dependent oxygenasesAnnual Review of Biochemistry 87:585–620.https://doi.org/10.1146/annurev-biochem-061516-044724
Article and author information
Author details
Publication history
- Version of Record published: September 12, 2019 (version 1)
Copyright
© 2019, Bersten and Peet
This article is distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use and redistribution provided that the original author and source are credited.
Metrics
-
- 1,266
- views
-
- 99
- downloads
-
- 6
- citations
Views, downloads and citations are aggregated across all versions of this paper published by eLife.
Download links
Downloads (link to download the article as PDF)
Open citations (links to open the citations from this article in various online reference manager services)
Cite this article (links to download the citations from this article in formats compatible with various reference manager tools)
Further reading
-
- Biochemistry and Chemical Biology
- Cell Biology
Hibernation is a period of metabolic suppression utilized by many small and large mammal species to survive during winter periods. As the underlying cellular and molecular mechanisms remain incompletely understood, our study aimed to determine whether skeletal muscle myosin and its metabolic efficiency undergo alterations during hibernation to optimize energy utilization. We isolated muscle fibers from small hibernators, Ictidomys tridecemlineatus and Eliomys quercinus and larger hibernators, Ursus arctos and Ursus americanus. We then conducted loaded Mant-ATP chase experiments alongside X-ray diffraction to measure resting myosin dynamics and its ATP demand. In parallel, we performed multiple proteomics analyses. Our results showed a preservation of myosin structure in U. arctos and U. americanus during hibernation, whilst in I. tridecemlineatus and E. quercinus, changes in myosin metabolic states during torpor unexpectedly led to higher levels in energy expenditure of type II, fast-twitch muscle fibers at ambient lab temperatures (20 °C). Upon repeating loaded Mant-ATP chase experiments at 8 °C (near the body temperature of torpid animals), we found that myosin ATP consumption in type II muscle fibers was reduced by 77–107% during torpor compared to active periods. Additionally, we observed Myh2 hyper-phosphorylation during torpor in I. tridecemilineatus, which was predicted to stabilize the myosin molecule. This may act as a potential molecular mechanism mitigating myosin-associated increases in skeletal muscle energy expenditure during periods of torpor in response to cold exposure. Altogether, we demonstrate that resting myosin is altered in hibernating mammals, contributing to significant changes to the ATP consumption of skeletal muscle. Additionally, we observe that it is further altered in response to cold exposure and highlight myosin as a potentially contributor to skeletal muscle non-shivering thermogenesis.
-
- Biochemistry and Chemical Biology
- Neuroscience
In most mammals, conspecific chemosensory communication relies on semiochemical release within complex bodily secretions and subsequent stimulus detection by the vomeronasal organ (VNO). Urine, a rich source of ethologically relevant chemosignals, conveys detailed information about sex, social hierarchy, health, and reproductive state, which becomes accessible to a conspecific via vomeronasal sampling. So far, however, numerous aspects of social chemosignaling along the vomeronasal pathway remain unclear. Moreover, since virtually all research on vomeronasal physiology is based on secretions derived from inbred laboratory mice, it remains uncertain whether such stimuli provide a true representation of potentially more relevant cues found in the wild. Here, we combine a robust low-noise VNO activity assay with comparative molecular profiling of sex- and strain-specific mouse urine samples from two inbred laboratory strains as well as from wild mice. With comprehensive molecular portraits of these secretions, VNO activity analysis now enables us to (i) assess whether and, if so, how much sex/strain-selective ‘raw’ chemical information in urine is accessible via vomeronasal sampling; (ii) identify which chemicals exhibit sufficient discriminatory power to signal an animal’s sex, strain, or both; (iii) determine the extent to which wild mouse secretions are unique; and (iv) analyze whether vomeronasal response profiles differ between strains. We report both sex- and, in particular, strain-selective VNO representations of chemical information. Within the urinary ‘secretome’, both volatile compounds and proteins exhibit sufficient discriminative power to provide sex- and strain-specific molecular fingerprints. While total protein amount is substantially enriched in male urine, females secrete a larger variety at overall comparatively low concentrations. Surprisingly, the molecular spectrum of wild mouse urine does not dramatically exceed that of inbred strains. Finally, vomeronasal response profiles differ between C57BL/6 and BALB/c animals, with particularly disparate representations of female semiochemicals.






