A quantitative modelling approach to zebrafish pigment pattern formation
Figures
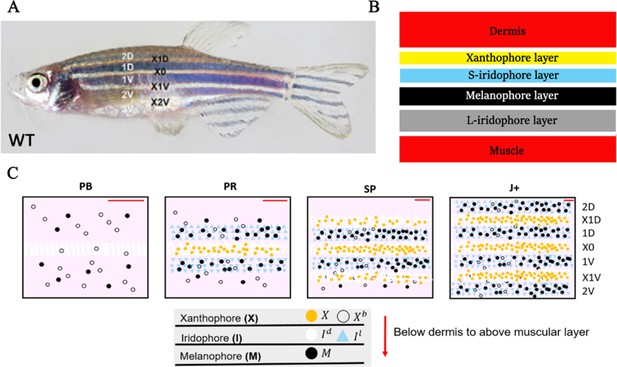
WT stripe composition and development.
(A) An adult wild type (WT) fish. Stripes and interstripes are labelled according to their order of temporal appearance. X0 is the first interstripe to appear. 1D and 1V (D - Dorsal, V- Ventral) are the first two stripes to appear. X1D and X1V are the next two interstripes to appear and so on. Image reproduced from Frohnhöfer et al., 2013 and licensed under CC-BY 4.0 (https://creativecommons.org/licenses/by/4.0). (B) Summary of pigment cell distribution in adult zebrafish. The cells in the xanthophore, S-iridophore, melanophore and L-iridophore layers consist of xanthophores and xanthoblasts, melanophores, S-iridophores and L-iridophores, respectively. Adapted from Hirata et al., 2003. (C) Schematic of WT patterns on the body of zebrafish. Stages PB, PR, SP, J+ correspond to developmental stages described in 3. Patterns form sequentially outward from the central interstripe, labelled X0, with additional dorsal stripes and interstripes labelled 1D, X1D, 2D, X2D from the centre (horizontal myoseptum) dorsally outward (similarly, ventral stripes and interstripes are labelled 1V, X1V, 2V, etc).
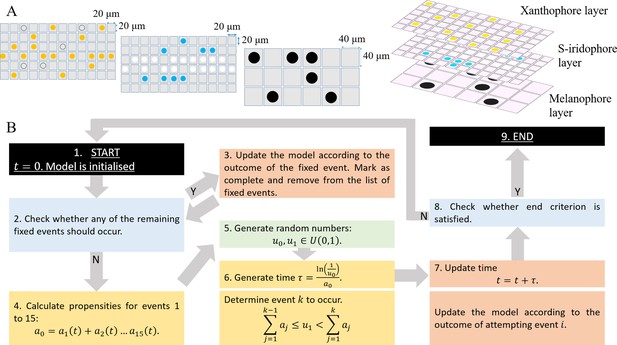
Model setup and simulation.
(A) An example model setup. The domain is made up of three layers. Layer which contains yellow (yellow circles) and unpigmented (clear circles with black outline). Layer which contains silvery (white circles) and blue (blue circles). Layer which contains black (black circles) only. Each lattice site on each of the respective layers contain at most one cell at any given time. The layers are stacked on top of each other as seen in real fish. We note that in our simulations the ordering of the layers does not play any role in determining pattern formation. (B) Schematics of model implementation. (1) The model is initialised as described in Appendix 1. (2) The model is checked for the requirements for a fixed event to occur (e.g. new cells differentiating at a given time). If there is a fixed event to be implemented, the algorithm moves to stage 3. Otherwise, the algorithm passes to stage 4. (3) The model is updated according to the fixed event and algorithm returns to stage 2. (4) The propensity functions - probabilities for all other events to occur are calculated. (5) Random numbers are generated. (6) Numbers are used to determine the time until the next event and which event will be implemented. (7) The model configuration is updated. (8) The algorithm checks if the end criterion is satisfied, that is in the case of WT, that . If so, the algorithm finishes. Otherwise the algorithm continues, returning to stage 2. (9) The simulation completes.
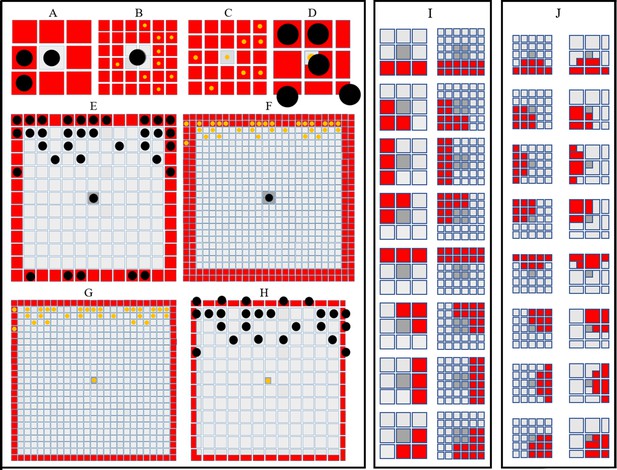
Simulating short- and long-range interactions.
(A–F) Comparing the number of cells in the short (A–D) and long (E–F) range distance (0.04 mm) from a central site, on different domain types. are represented as black circles. are represented as yellow circles. (A–B) A visualisation of sites (marked in red) local to central melanocyte on the (A) melanocyte domain and (B) xanthophore domain.(C–D) A visualisation of sites (marked in red) local to central xanthophore on the (C) xanthophore domain and (D) melanocyte domain. (E–F) A visualisation of sites (marked in red) long-distance to central melanocyte on the (E) melanocyte domain and (F) xanthophore domain.(G–H) A visualisation of sites (marked in red) long-distance to central xanthophore on the (G) xanthophore domain and (H) melanocyte domain. (I) Melanophore movement with respect to local neighbours. If a melanophore located in a central position (marked in dark grey) attempts to move, the melanophore will consider neighbours in all sites marked in red on the melanophore domain (left), xanthophore and S-iridophore domain (right). (J) Movement on xanthophore/iridophore domain with respect to local neighbours. If a xanthophore/iridophore located in a central position (marked in dark grey) attempts to move, the xanthophore/iridophore will consider neighbours in all sites marked in red on the xanthophore and S-iridophore domain (left), melanophore domain (right).
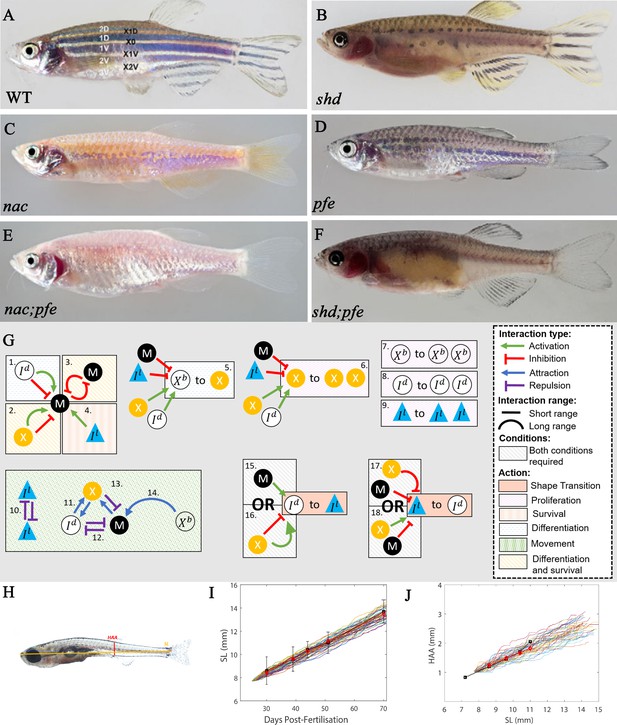
‘Missing cell’ mutants and cell-cell interactions implemented.
(A–F) Adult phenotype of the set of fish used to deduce S-iridophore interactions. (A) WT fish, (B) shady (shd), (C) nacre(nac), (D) pfeffer (pfe), (E) nac;pfe, (F) shd;pfe. (G) A representation of all the interactions implemented in the model. See Supplementary files 4 and 5 for a detailed justification of the rates and interaction types, respectively. See Supplementary file 6 for a detailed justification of all parameters. The direction of the arrow/inhibition sign indicates which cell is acting on which. For example in assumption 13, the blue arrow from melanophore to xanthophore indicates that melanophores attract (green background) xanthophores. The purple inhibition arrow indicates that the xanthophores repel melanophores. OR statements in assumptions 15–18 are logical OR statements. For example, conditons 15 and 16 indicate that a dense S-iridophore may become loose if there are melanophores in the short range OR, there are no xanthophores in the short range AND many xanthophores in the long range. (H) A visualisation of measurements HAA and SL. (I) Plots of SL versus days post-fertilisation (dpf) for 40 model realisations. Each coloured line is a single realisation. Black squares are SL versus days post-fertilisation extracted from Parichy et al., 2009 in real fish given in Supplementary file 3. Red diamonds are the mean SL versus days post-fertilisation from 40 simulations. (J) Plots of SL (mm) versus HAA (mm) for 40 model realisations. Each coloured line is a single realisation. Black squares are SL versus HAA extracted from Parichy et al., 2009 given in Supplementary file 3. Red diamonds are the mean HAA versus SL from 40 simulations. Error bars are one standard deviation. The model agrees well with the data in both cases. Images (A–F) reproduced from Frohnhöfer et al., 2013 and licensed under CC-BY 4.0 (https://creativecommons.org/licenses/by/4.0).
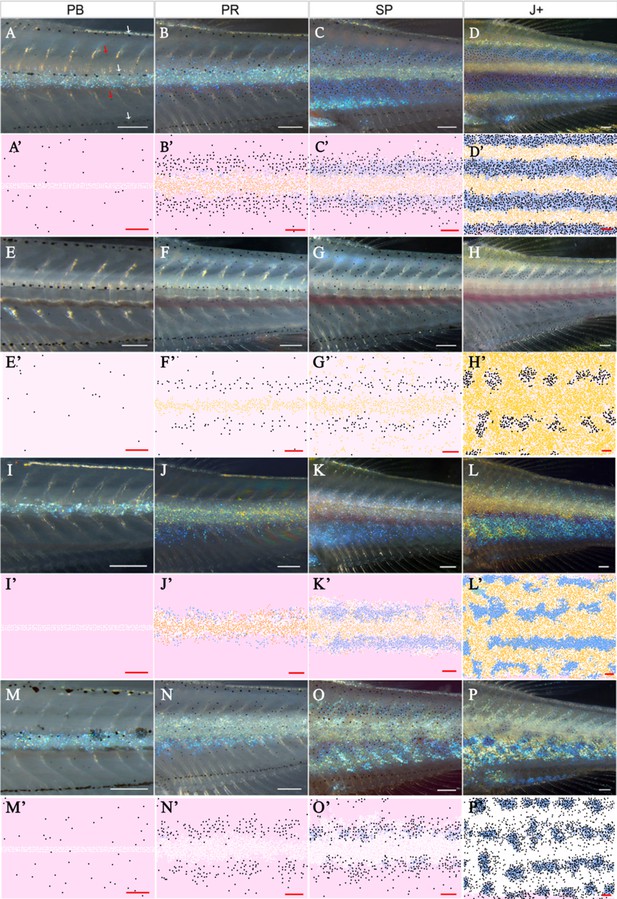
Representative simulation of WT, shd, nac and pfe.
(A–D), (E–H), (I–L), (M–P) WT, shd, nac, pfe development respectively and (A’–D’), (E’–H’), (I’–L’), (M’–P’) corresponding model simulation. Red arrows: melanocytes in the skin layer, modelled in (A). White arrows indicate the embryonic pattern of melanocytes in four stripes, that are deeper than the skin level and are consequently not included in the model. See Videos 1–4 for representative examples of these simulations in movie format. Scale bar is 0.25 mm for all images. Experimental images (A–D), (E–H), (I–L), (M–P) reproduced from Frohnhöfer et al., 2013 and licensed under CC-BY 4.0 (https://creativecommons.org/licenses/by/4.0).

Example WT simulations at stage J+ when the parameters governing the rate of proliferation, movement, differentiation and death are perturbed slightly.
Each square is an example WT simulation at stage J+ where each rate parameter is perturbed to 1+ times its normal value. The value is chosen uniformly at random from the set .
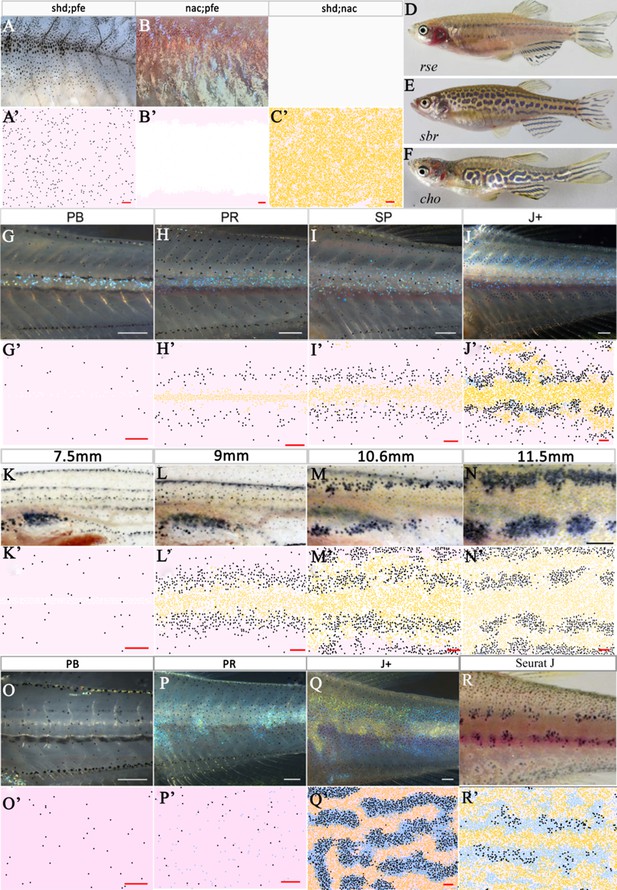
Other mutant fish simulations.
(A–C) Real stage J+ shd;pfe, nac;pfe, shd;nac mutants, respectively. (A’–C’) Model simulations at stage J+ for shd;pfe, nac;pfe, shd;nac mutants, respectively. (D–F) Adult phenotype of selected (D) rse, (E) sbr, (F) cho mutant. (G–J), (K–N), (O–Q) rse, sbr and cho development, respectively. (G’–J’), (K’–N’), (O’–Q’) rse, sbr and cho model simulations, respectively. (R) seurat at stage J. (R’) seurat model simulation at stage J. Scale bar is 0.25 mm for all images. See Videos 5–7 for representative examples of these simulation in movie format. Experimental images (A–C), (D), (F) (G–J) and (O–Q) are reproduced from Frohnhöfer et al., 2013, (E), (K–N) from Fadeev et al., 2015, (R) from Eom et al., 2012 and are all licensed under CC-BY 4.0 (https://creativecommons.org/licenses/by/4.0).
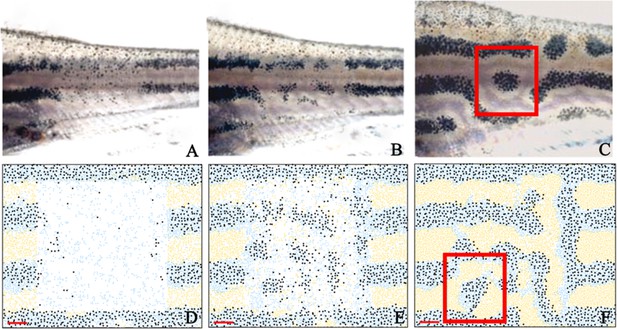
Stripe regeneration simulations.
(A–C) Regeneration of new pigment producing cells 7, 14 and 21 days respectively after a small rectangular window of cells in the adult WT stripes are completely ablated (Patterson and Parichy, 2013). (D–F) A representative simulation of the regeneration of an adult zebrafish 7, 14 and 21 days after a simulated ablation has occurred. Scale bar is 0.25 mm in all images. Experimental images (A–C) are reproduced from Yamaguchi et al., 2007. Copyright (2007) National Academy of Sciences U.S.A.; these images not covered by the CC-BY 4.0 licence and further reproduction of this panel would need permission from the copyright holder.
© 2007 National Academy of Sciences USA. Experimental images (A–C) are reproduced from Yamaguchi et al., 2007, with permission of PNAS. These images not covered by the CC-BY 4.0 licence and further reproduction of this panel would need permission from the copyright holder.
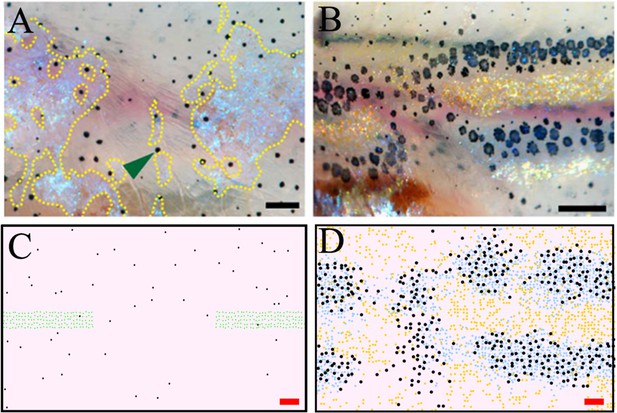
Stripe regeneration post S-iridophore ablation.
(A) S-iridophores are ablated using pnp4a:NTR+Mtz at stage PB (B) Stripe development in wild-type fish following S-iridophore ablation. Xanthophores are associated with S-iridophore interstripe. Melanophores aggregate around the new interstripe. (C–D) A representative simulation of S-iridophore ablation and subsequent development. For clarity, dense S-iridophores are displayed in green in (C). Scale bar is 0.25 mm in all images. Experimental images (A–B) are reproduced from Patterson and Parichy, 2013 and licensed under CC-BY 4.0 (http://creativecommons.org/licenses/by/4.0).
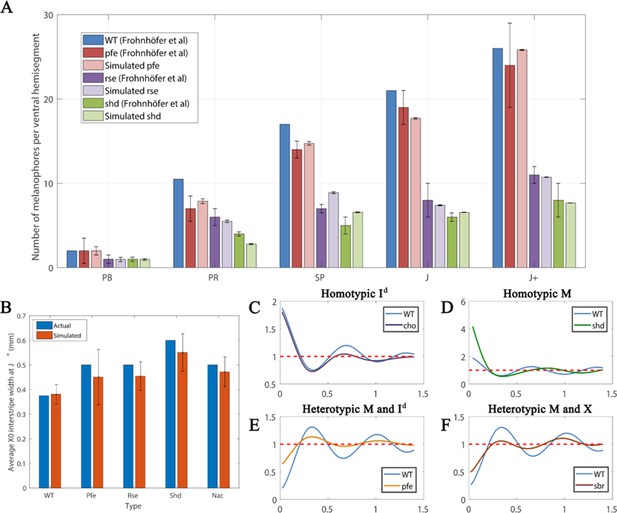
Quantitative analyses.
(A) Number of melanocytes per ventral hemisegment. Quantification of melanocytes. A comparison of melanocyte numbers between mutants and WT seen in our simulations and data recreated from Frohnhöfer et al., 2013. For each stage, our simulated number of melanocytes was normalised by the WT number in data by Frohnhöfer et al., 2013. (The number of animals used for counting was at least five for each measurement point.) The number of simulations was 100. Error bars indicate the standard deviation. There is no error bar for WT, since WT data was used to normalise the data. (B) X0 interstripe width. A comparison of X0 stripe width in 100 simulations with X0 stripe width in real mutants (taken from representative images; Frohnhöfer et al., 2013). The darker bars depict experimental data, lighter bars depict our simulated data. Error bars indicate the standard deviation. Since there is only one image for each of the respective mutants, there is no error bar for actual (real experimental data). (C–F) PCF for different cell types averaged over 100 simulations using the square uniform PCF (Gavagnin et al., 2018). The spatial pair correlation as a function of distance for (C) dense S-iridophores only (D) melanocytes only, (E) melanocytes and dense S-iridophores, (F) melanocytes and xanthophores. Experimental data in (A) is reproduced from Frohnhöfer et al., 2013.
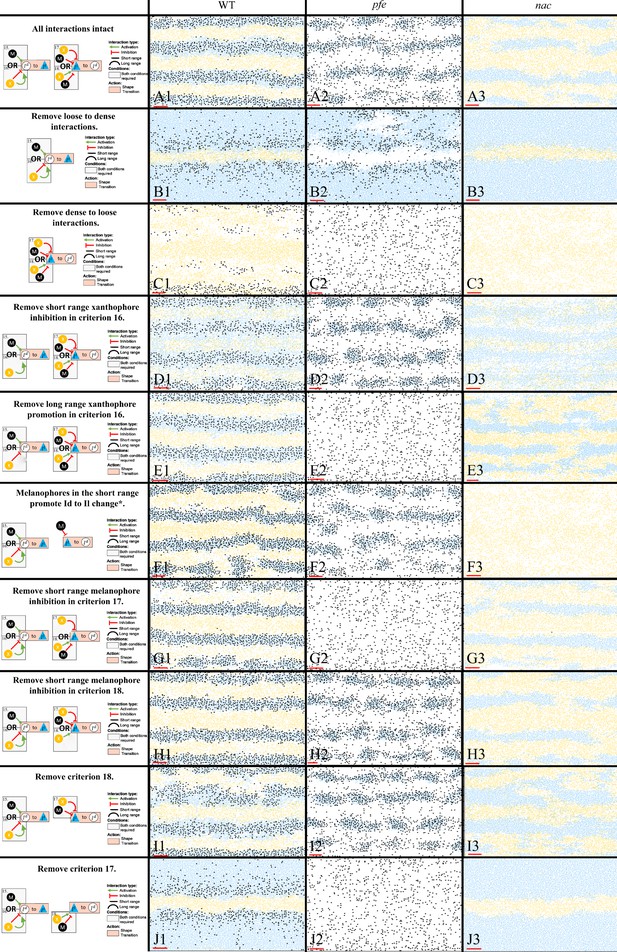
Representative simulations of the model with some of the S-iridophore interactions removed.
The first column displays a diagram of the S-iridophore interactions which remain (all other cell-cell interactions are unchanged). Columns 2–4 are representative simulations of WT, pfe and nac under these conditions. *This interaction is equivalent to removal of long range xanthophore inhibition in criterion 17. It is also equivalent to removal of short range xanthophore promotion in criterion 18.
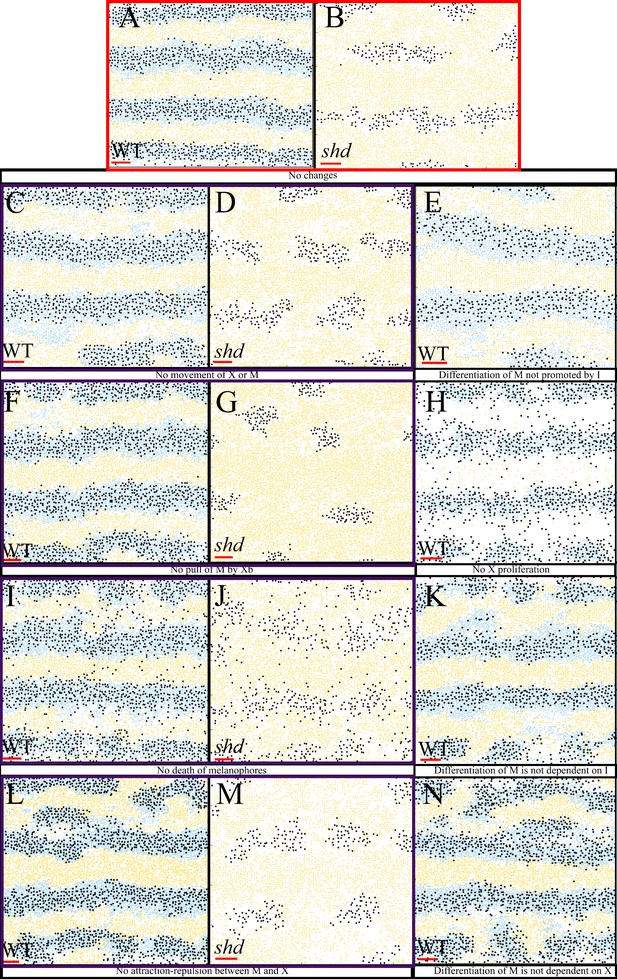
Representative simulation of the model with certain rules omitted.
(A–B) WT and shd respectively with all rules included. (C–D) No movement of melanocytes or xanthophores for WT and shd, respectively. (E) Differentiation of melanocytes only promoted by dense S-iridophores for WT fish that is the rate of melanocytes does not depend on the number of S-iridophores. (F–G) No pull of melanocytes by xanthoblasts for WT and shd fish, respectively. (H) No proliferation of xanthophores for WT. (I–J) No melanocyte death for WT and shd fish. (K) Differentiation of melanocytes is not dependent on S-iridophores, (L–M) No death of melanocytes for WT and shd (N) Differentiation of melanocytes is dependent on dense S-iridophores being present in the long distance only.
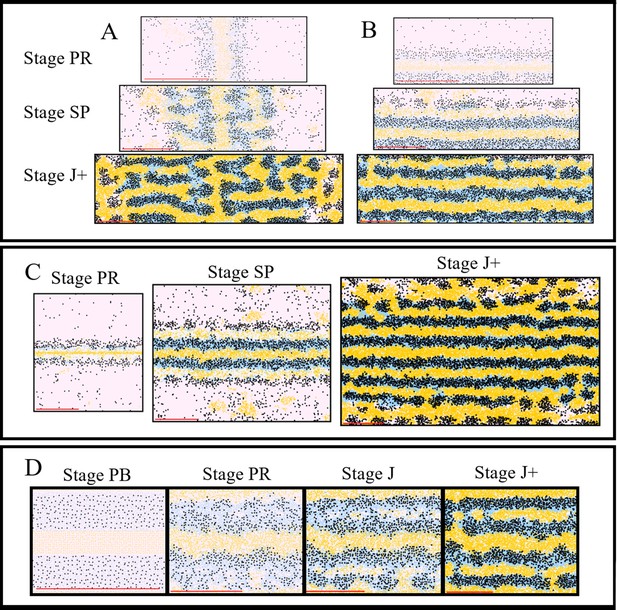
In silico investigation into important mechanisms for controlling pattern formation.
The simulated domains at stages PR, SP and J+ wherein the following are changed (A) the orientation of the initial S-iridophore interstripe, (B) the initial position of the S-iridophore interstripe, (C) the initial domain size (D) the initial domain so that it is populated with adult-width stripes and interstripes.
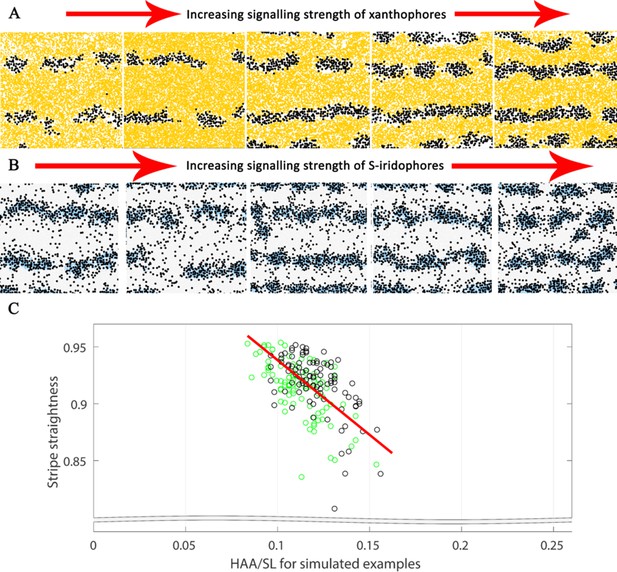
Substantial alterations in starting conditions generate major alterations in pattern formation.
The effects of different signalling strengths in pfe and shd. Representative images of J+ simulations of (A) shd and (B) pfe where the signalling strength of melanocyte differentiation in the long range is increased (from left to right). Increasing the signalling strength of the respective cell types decreases the width of the interstripe X0 and increases the number of pseudo-stripes from 2 to 4. For our simulations, we choose the weakest shown signalling strength for xanthophores, corresponding to the furthest left of (A) and the strongest shown signalling strength for S-iridophores, corresponding to the furthest right of (B). (C) Stripe straightness of our simulations correlates with the simulated HAA/SL ratio. Each circle is one simulation, green circles indicate simulations up to stage J, black circles indicate simulations of up to stage J+. There are one hundred simulations in total for each stage. The red line is the line of best fit.
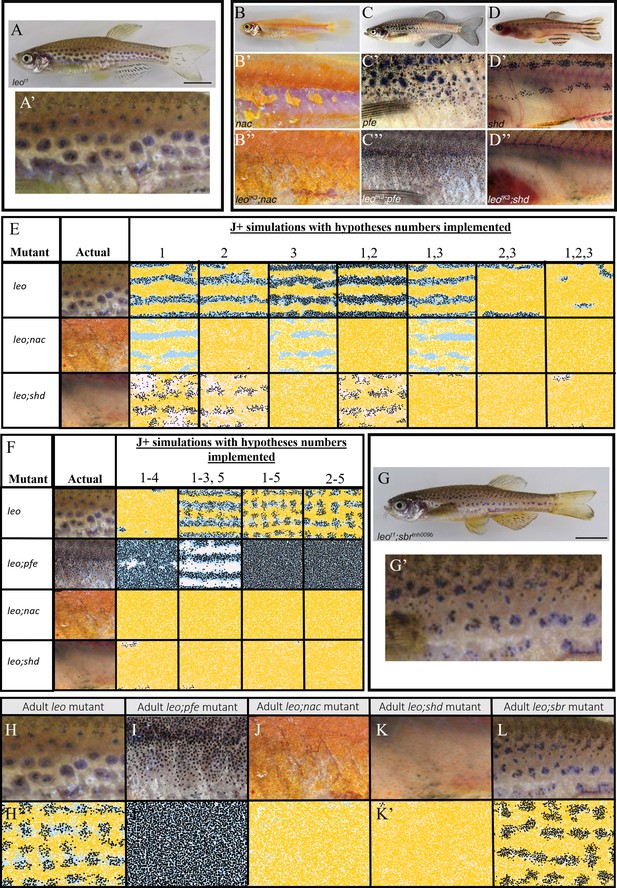
Using the model to generate predictions regarding the underlying effects of the leo mutant.
(A–D) Adult leo, nac, pfe and shd respectively. (A’–D’) The flanks of leo, nac, pfe and shd respectively. (B’’–D’’) The flanks of leo;nac, leo;pfe and leo;shd respectively. (E) Example simulations where combinations of hypotheses 1–3 in the text are implemented. (F) Example simulations where combinations of hypotheses 1–5 in the text are implemented. Note that simulations of 1–5 are similar to simulations for 2–5, indicating that hypotheses one is not necessary to predict the leo phenotype. (G) Adult leo;sbr (G’) Flank of leo;sbr. (H–L) Adult mutants contrasting with (H’–L’) simulations using hypotheses 1–5 from the main text. (H–L) Flank of leo, leo;nac, leo;pfe, leo;shd and leo;sbr respectively. (H’–L’) Simulated images of leo, leo;pfe, leo;nac, leo;shd and leo;sbr respectively at stage J+. (A), (A’), (G), (G’), (H) and (L) from Fadeev et al., 2015, (B–D’’), (I–K) from Irion et al., 2014 and are all licensed under CC-BY 4.0 (http://creativecommons.org/licenses/by/4.0).
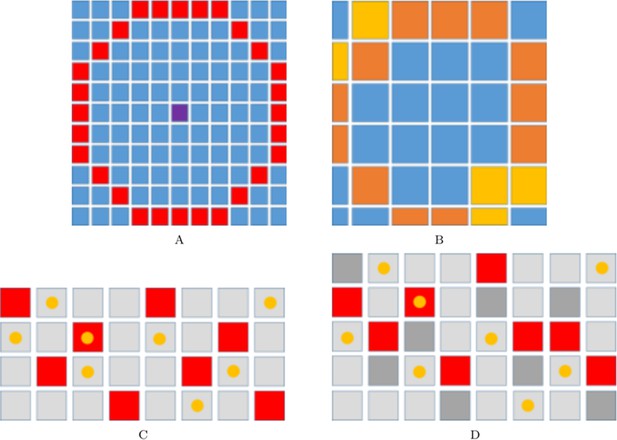
Implementation of xanthoblast-melanocyte communication and domain growth.
(A–B) Range for xanthoblast cytonemes in our model. Figure:range_pull The range of xanthoblast cytonemes on . The centre lattice site (marked in purple) represents site . The lattice sites labelled in red are those sites that lie in the range of the cytonemes of an occupying site (the sites given by set ). Figure:range_pull_mel The range of xanthoblast cytonemes on . The sites indicated with in orange or yellow are those sites that can be reached by the range of in . The colour of any given site reflects the number of corresponding sites on , , , that are within range of the cytonemes (marked in red in (A)) and thus the probability of the site being chosen in relation to other coloured sites. Specifically, sites labelled in orange are twice as likely to be chosen than those sites marked in yellow. (C–D) An example growth event in the vertical direction. Squares represent sites in a given domain. Yellow circles represent cells occupying these sites. (C) First a site is chosen at random from each column (marked in red). (D) Next a new site is inserted either above or below the first site with probability . Each of the chosen sites are marked in dark grey.

Example shd, nac, pfe and leo simulations at stage J+ when the parameters governing the rate of proliferation, movement, differentiation and death are perturbed.
Each rectangle is an example simulation at stage J+ for which each rate parameter is perturbed to 1+ times its normal value. The value is chosen uniformly at random from the interval .
Videos
Simulated development of WT.
Simulated development of shd.
Simulated development of nac.
Simulated development of pfe.
Simulated development of rse.
Simulated development of sbr.
Simulated development of cho.
Additional files
-
Supplementary file 1
Summary of all notation used in Appendix 1.
- https://cdn.elifesciences.org/articles/52998/elife-52998-supp1-v1.pdf
-
Supplementary file 2
Summary of all notation used in Appendix 1 regarding short and long range interactions.
- https://cdn.elifesciences.org/articles/52998/elife-52998-supp2-v1.pdf
-
Supplementary file 3
SL measurements and HAA measurements and WT pattern description by stage are given as by Parichy et al., 2009.
Corresponding dpf for the stages are approximated from the images of Frohnhöfer et al., 2013.
- https://cdn.elifesciences.org/articles/52998/elife-52998-supp3-v1.pdf
-
Supplementary file 4
All continuous time events and their corresponding propensities for event attempts at time , denoted for event in minutes.
- https://cdn.elifesciences.org/articles/52998/elife-52998-supp4-v1.pdf
-
Supplementary file 5
Full description of all the cell–cell interaction given in main text.
Melanocytes, xanthophores, loose iridophores, dense iridophores and xanthoblasts are denoted , , , , respectively. C1, C2 stands for cell 1, cell 2, where , is the target cell and cell two is the signalling cell with corresponding signal range (R) that is either short (S), up to 0.04 mm, or long (L), 0.12 mm. This signal generates action (A) of cell type C1 of: movement (M), differentiation (D), proliferation (P) or survival (S) of type (T). For action (D) or (S), type (T) is denoted ‘+’ if the resultant action is promotion of action A and ‘-’ if the resultant action is inhibition of action (A). For action (M), type (T) is denoted ‘+’ if the resultant action is attraction towards cell type C1 and ‘-’ if the resultant action is repulsion away from cell type C1. *Melanocytes can also differentiate randomly, independent of any other cell type.
- https://cdn.elifesciences.org/articles/52998/elife-52998-supp5-v1.pdf
-
Supplementary file 6
Parameters implemented in the model.
- https://cdn.elifesciences.org/articles/52998/elife-52998-supp6-v1.pdf
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/52998/elife-52998-transrepform-v1.pdf