Conformational changes in twitchin kinase in vivo revealed by FRET imaging of freely moving C. elegans
Figures
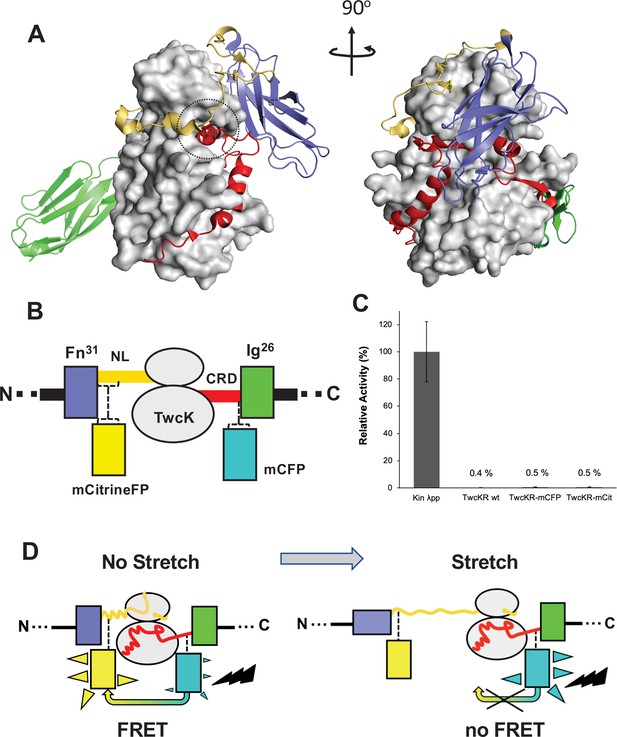
Molecular composition of the TwcK-FRET sensor.
(A) Crystal structure of the multi-domain, autoinhibited TwcK (PDB code 3UTO). The ATP pocket, blocked by the CRD extension, is marked with a circle. Domain color code as described in (B). (B) Domain composition of the TwcK region, illustrating the sites of mCitrineFP and mCFP incorporation. (C) Activity measurements of multi-domain TwcK variants using the Kinase Glo luminescent assay (Kin λpp) indicate the twitchin kinase domain that displays maximal activity by lacking flanking tails and having been treated with λphosphatase (Williams et al., 2018). (D) Functional principle of the TwcK-FRET stretch-sensing construction. The non-stretched state represents the basal molecular state, where NL and CRD tail extensions pack against the kinase. Unfolding and extension of the NL segment (and/or CRD sequence) are expected to happen concurrently and result in a stretched state.
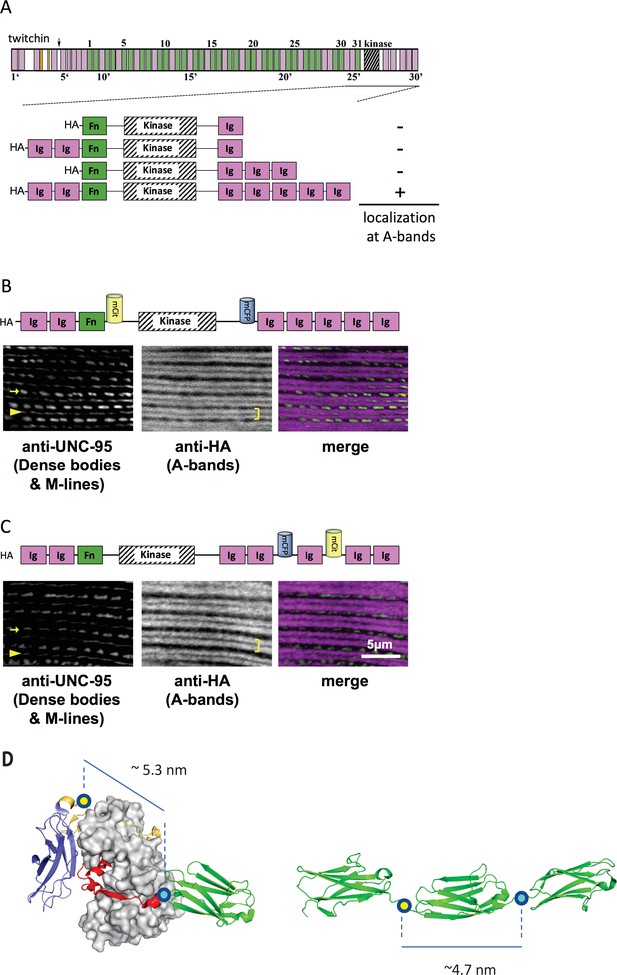
A portion of twitchin containing FRET moieties properly localizes in the sarcomere.
(A) Schematic representation of domain organization of twitchin and segments containing the kinase domain tested for localization to A-bands. Note that HA-Ig-Ig-Fn-NL-Kin-CRD-Ig-Ig-Ig-Ig-Ig localizes to sarcomeric A-bands, the normal location of full-length twitchin. (B) The minimally localizing twitchin fragment containing FRET moieties surrounding the kinase domain localizes to A-bands. Above, schematic; below, immunofluorescent localization. (C) The minimally localizing twtichin fragment containing FRET moieties surrounding Ig28 also localizes to A-bands. Above, schematic; below, immunofluorescent localization. Arrow, row of dense bodies; arrowhead, M-line; bracket, A-band. (D) Structural representations of the test (left) and control (right) molecular loci, including estimated distances between the sites of FP incorporation. The test molecule is the crystal structure of the multi-domain twitchin kinase (PDB 3UTO). The control molecule is a model of the Ig27-Ig28-Ig29 segment based on available structural data for homologous poly-Ig tandems of human titin (von Castelmur et al., 2008).
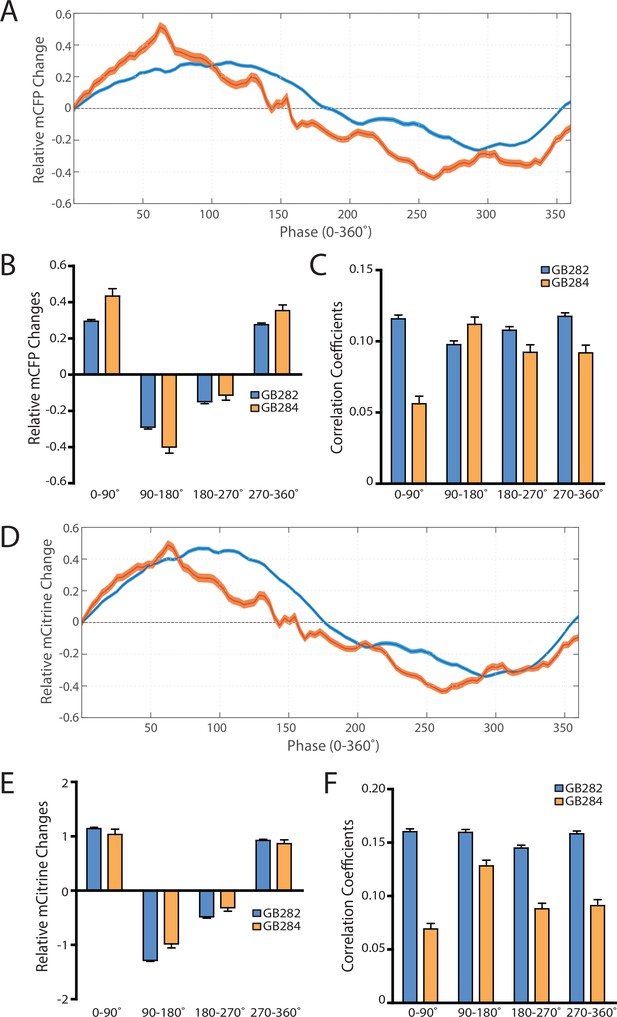
As controls for artifacts, analysis on individual fluorophores showing the absence of consistent increases in change and correlations in all parts of the contraction cycle, comparing GB282 to GB284.
(A) Average mCFP intensity changes for GB282 and GB284 aligned to same cycles of muscle contractions as in Figure 7A. (B) Average mCFP intensity value changes for GB282 and GB284 for each part of a muscle contraction cycle (n≥5953 cycles, 30 animals). (C) Average correlation coefficients between mCFP intensity value changes and curvature for GB282 and GB284 for each part of a muscle contraction cycle (n≥5953 cycles, 30 animals). (D) Average mCitrine intensity changes for GB282 and GB284 aligned to same cycles of muscle contractions as in Figure 7A. (E) Average mCitrine intensity value changes for GB282 and GB284 for each part of a muscle contraction cycle (n≥5953 cycles, 30 animals). (F) Average correlation coefficients between mCitrine intensity value changes and curvature for GB282 and GB284 for each part of a muscle contraction cycle (n≥5953 cycles, 30 animals).
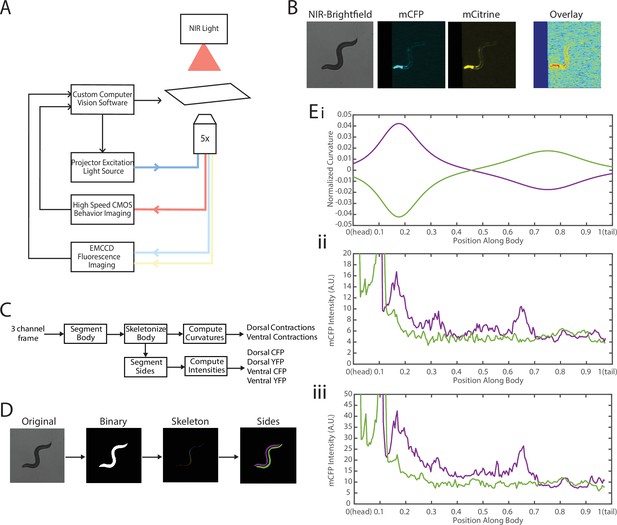
Experimental platform for posture tracking and fluorescence imaging.
(A) Schematic of the platform adapted from Stirman et al., 2011, which uses an inverted microscope with a 5× objective with two optical channels to image in brightfield using NIR and fluorescence imaging using a projector to provide excitation light (see Materials and methods). Tracking is performed by live analysis of a subsample of images, maintaining the pharynx of the animal in the center of the FOV of fluorescence images (see Materials and methods). (B) Sample frames acquired using the platform, showing the three channels captured at each time point: NIR brightfield (left), mCFP (center), and mCitrineFP (right). (C) The three channels are spatially aligned to allow for accurate extraction and comparisons of muscle contractions and FRET values. (D) Schematic of analysis pipeline. The algorithm is performed on each frame, taking as inputs the raw three-channel images. The brightfield image is processed to produce a binary mask of the body outline of the animal. The binary image is then used to create an ordered set of points along the midline of the animal, from head to tail. The midline and body outline are subsequently used to create individual binary mask images of the dorsal and ventral sides of the animal. A tracking algorithm is used to robustly characterize the two sides separately (blue and red). (E) Using the midline of the animal, (i) the magnitude of contractions are characterized as computed curvature values, and using the mask images, (ii, iii) the fluorescence intensities are characterized along the length of the animal for both the dorsal and ventral sides (0=head, 1=tail) (green and purple traces represent the two sides). FOV, field of view; NIR, near-infrared.
Tracking of individual sides throughout recordings was performed by comparing angles between three vectors.
The centroid of each side mask to the closest point in the midline (blue and orange), and the tail to head vector using the midline endpoints (black).
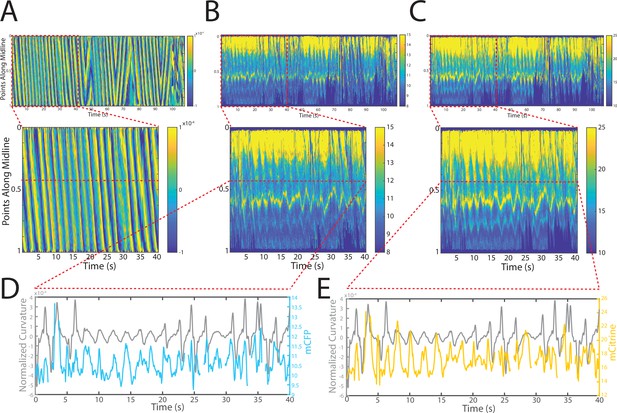
Tracking platform and analysis pipeline provide precise spatio-temporal measurements of normalized curvature and fluorescence intensities.
(A) Kymograph of sample measurements of curvature (C(x,t)) for one side during a recording, where y-axis represents the position along the anterior-posterior axis of the animal, and x-axis represents time. Colormap indicates the computed curvature value. (B, C) Kymographs (B) of mCFP and mCitrine fluorescence measurements (C) in the same side for the recording in (A), using the same y-axis and x-axis. Colormap indicates the quantified fluorescence emission. (D, E) Sample traces of normalized curvatures (black), and mCFP (blue) in (D), and mCitrine (yellow) in (E), intensities for a given point along the animal.
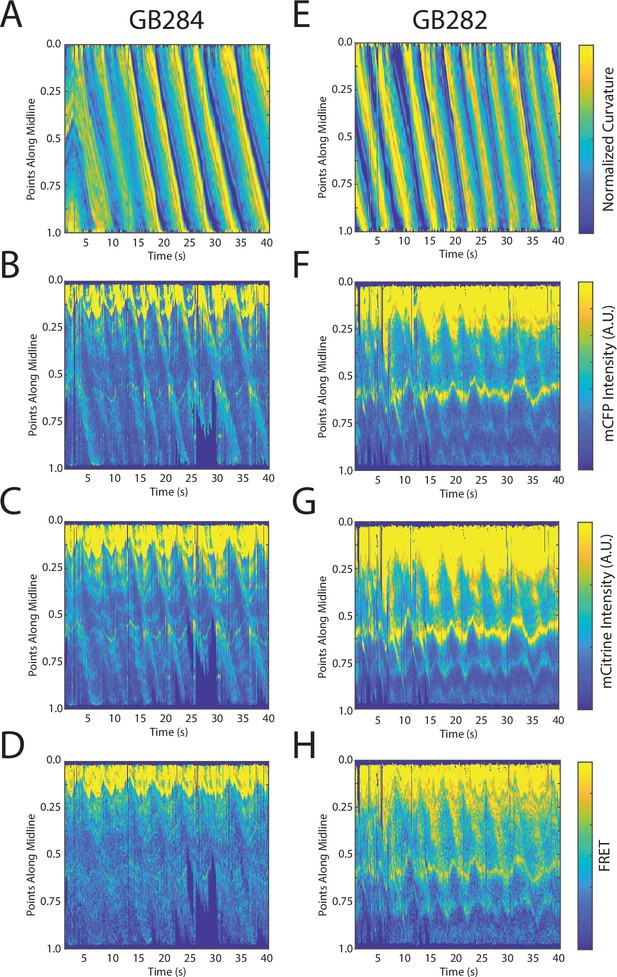
FRET signals for GB282 and GB284 are qualitatively correlated with muscle contractions.
(A–D) Sample measurements for GB284 of curvature (A), mCFP (B), mCitrine (C), and FRET (D) of a recording for a representative worm. (E–H) Sample measurements for GB282 of curvature (E), mCFP (F), mCitrine (G), and FRET (H) of a recording for a representative worm. Fluorescence intensities and FRET quantifications are normalized as described in Materials and methods.
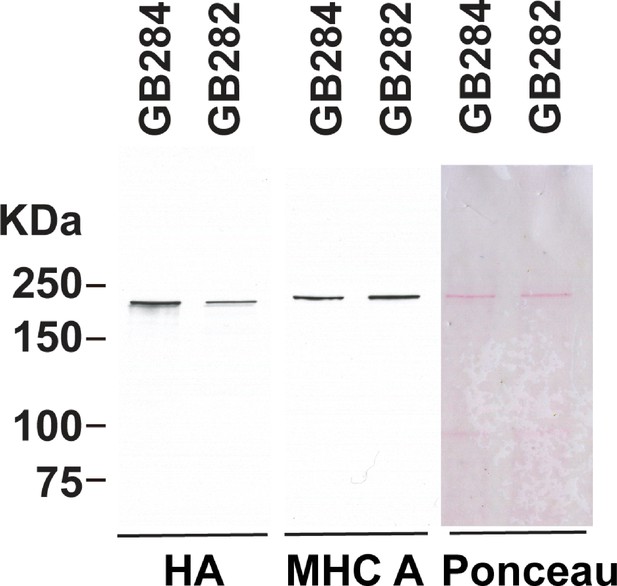
Higher level of expression of the fusion protein from the control GB284 as compared with the tester GB282.
GB282 and GB284 express the same portion of twitchin (kinase domain and flanking domains) but with different placements of the FRET moieties (Figure 2), and the transgenic arrays are integrated into the genome. Total protein extracts were prepared from each strain, run out on SDS-PAGE, transferred to nitrocellulose, and reacted against either antibodies to myosin heavy chain A (MHC A) or to HA (for detection of the fusion protein). From right to left are shown Ponceau staining of the blot, and reaction versus antibodies to MHC A or HA. The positions of molecular weight size markers are indicated. The experiment was repeated four times and a mean value of 2.10-fold higher expression from GB284 versus GB282 was found.
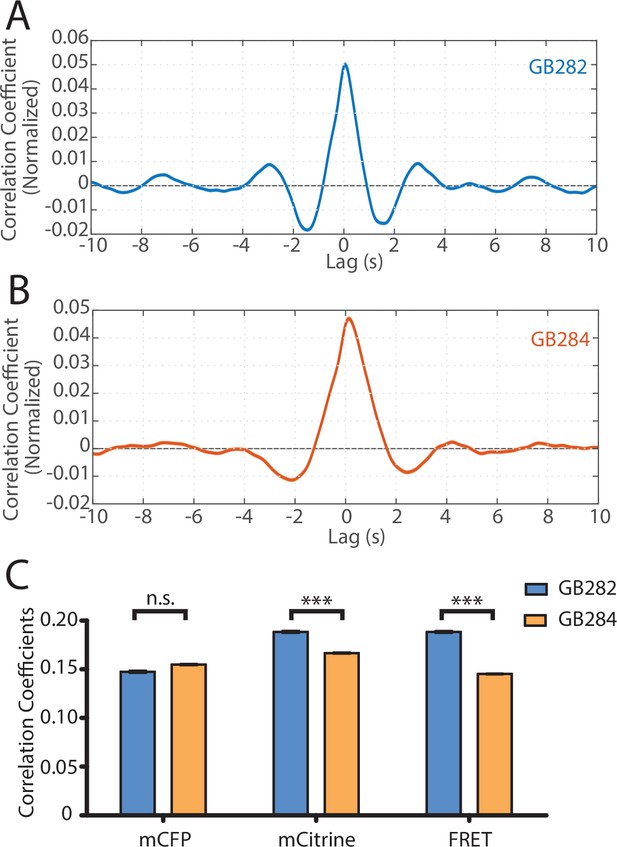
Statistical analysis reveals correlation between muscle contractions and FRET activity.
(A) Cross-correlation analysis between quantified curvature and FRET values for GB282 (n=27,677 midline points, 30 animals). (B) Cross-correlation analysis between quantified curvature and FRET values for GB284 (n=29,558 midline points, 30 animals). (C) Correlation coefficients between muscle contractions and mCFP, mCitrine, and FRET for strains GB282 (n=27,677 midline points, 30 animals) and GB284 (n=29,558 midline points, 30 animals) (Student’s t-test).
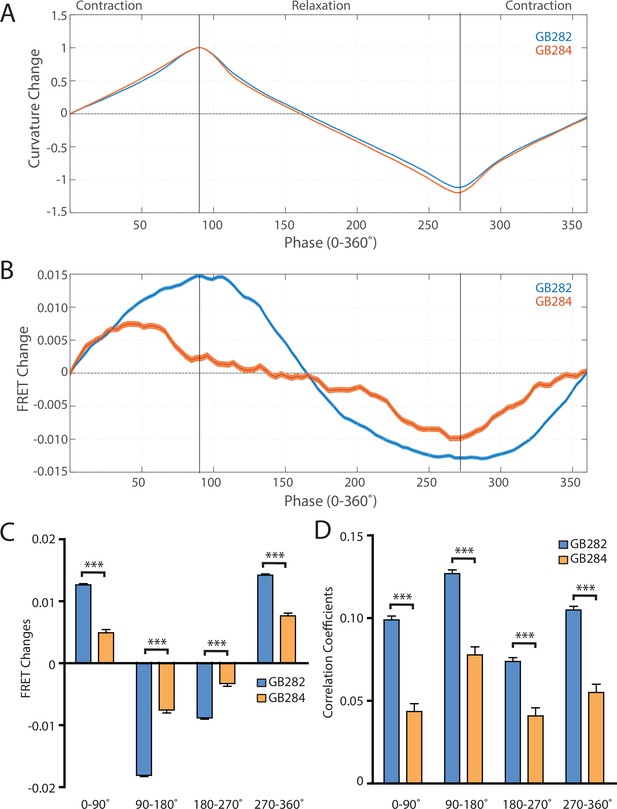
Changes in FRET during muscle contraction cycles suggest a conformational change.
(A) Average normalized curvature values aligned to cycles of muscle contractions, with 90° representing maximum contraction, and 270° representing maximum relaxation, for GB282 (blue, n≥13,840 cycles, 30 animals) and GB284 (orange,n≥11,707 cycles, 30 animals). (B) Average FRET value changes for GB282 (blue) and GB284 (orange) aligned to same cycles of muscle contractions as in (A). Note that FRET diminishes during muscle relaxation (90–270°). (C) Average FRET value changes for GB282 and GB284 for each part of a muscle contraction cycle. Statistical comparisons of changes in FRET show statistically higher changes in magnitude for GB282 in comparison to GB284 (Student’s t-test, n≥11,707 cycles, 30 animals). ***p≤0.001. (D) Average correlation coefficients between FRET value changes and curvature for GB282 and GB284 for each part of a muscle contraction cycle. Statistical comparisons of changes in FRET show statistically higher correlation coefficients for GB282 in comparison to GB284 (Student’s t-test, n≥11,707 cycles, 30 animals). ***p≤0.001. All normalized curvature and FRET values were quantified as described in Materials and methods.
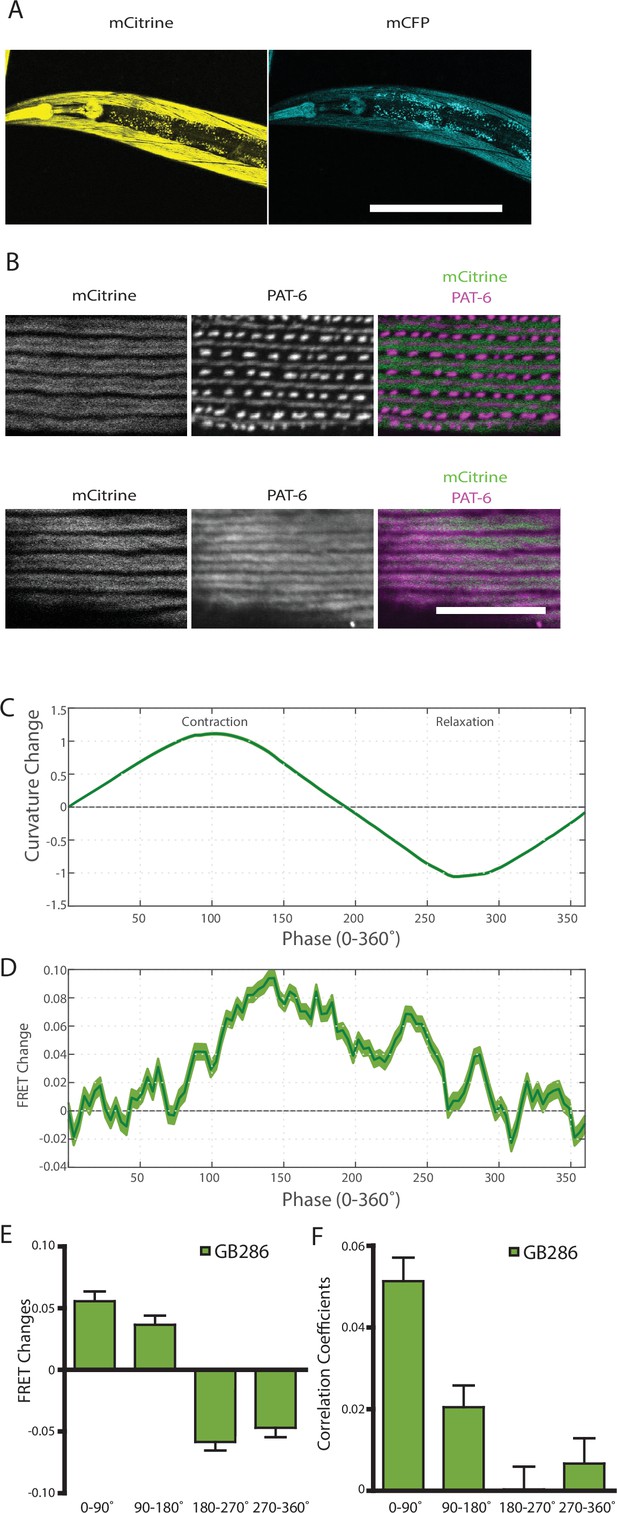
Nematodes expressing full-length twitchin with FRET moieties around the kinase domain (GB287) phenocopies the transgenic line (GB282).
(A) CRISPR/Cas9 gene-edited strain (GB286) expresses endogenous full-length twitchin with FRET moieties in the appropriate cells (body wall and pharyngeal muscle). This twitchin fusion can be imaged either for the mCitrine or mCFP moiety. Scale bar, 100 μm. (B) The twitchin FRET fusion protein localizes to the proper location in sarcomeres, A-bands. The top row shows co-imaging of the mCitrine signal with immunostaining for PAT-6 (α-parvin), a marker for the base of M-lines and dense bodies (Z-disks). The bottom row show co-localization of the mCitrine signal and immunostaining for twitchin. Scale bar, 10 μm. (C) Average normalized curvature values aligned to cycles of muscle contractions, with 90° representing maximum contraction, and 270° representing maximum relaxation, for GB287 (n≥5953 cycles, 30 animals). (D) Average FRET value changes for GB287 aligned to same cycles of muscle contractions as in (C). (E) Average FRET value changes for GB287 for each part of a muscle contraction cycle (n≥5953 cycles, 30 animals). (F) Average correlation coefficients between FRET value changes and curvature for GB287 for each part of a muscle contraction cycle (n≥5953 cycles, 30 animals).
Videos
A freely moving worm being tracked and imaged in dual modality.
Two-camera microscopy system enables simultaneous single-animal tracking in bright-field mode (left) and FRET measurement in fluorescent mode (right—mCFP and mCitrine).
Additional files
-
Supplementary file 1
DNA sequences of the primers used in this work.
- https://cdn.elifesciences.org/articles/66862/elife-66862-supp1-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/66862/elife-66862-transrepform1-v2.docx






