Legionella pneumophila regulates host cell motility by targeting Phldb2 with a 14-3-3ζ-dependent protease effector
Figures
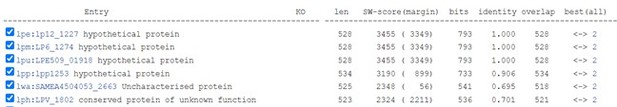
Lem8 is a cysteine protease-like Dot/Icm effector toxic to yeast.
(A) Alignment of Lem8 with several known cysteine proteases obtained by PSI-BLAST analysis. The strictly conserved catalytic residues are marked in red. Shown cysteine proteases are HopN1 and AvrpphB from P. syringae, YopT from Y. enterocolitica, and PfhB1 from P. multocida. (B) Lem8 is translocated into mammalian cells via the Dot/Icm transporter. U937 cells were infected with wild-type L. pneumophila or the dotA− mutant expressing the β-lactamase-Lem8 fusion. One hour after infection, the CCF4-AM fluorescence substrate was added into the cultures and the cells were incubated for another 2 hr at room temperature before image acquisition. Cells emitting blue fluorescence signals were quantitated by counting at least 500 cells in each experiment done in triplicate. Results shown are mean ± standard error (SE) from one representative experiment. (C) Expression profile of lem8 in L. pneumophila grown in AYE broth supplemented with thymidine. Bacteria grown to stationary phase were diluted at 1:20 in fresh medium and subcultures were grown in a shaker. Bacterial growth was monitored by measuring OD600 at the indicated time points. Equal amounts of bacterial cells were lysed for measurement of Lem8 levels by immunoblotting with Lem8-specific antibodies. The metabolic protein isocitrate dehydrogenase (ICDH) was probed as loading control. (D) Lem8 is toxic to yeast in a manner that requires the predicted Cys-His-Asp motif. Yeast strains expressing Lem8 or the indicated mutants from the galactose-inducible promotor were serially diluted and spotted on the indicated media. The plates were incubated at 30°C for 48 hr before image acquisition. The expression of Lem8 and its mutants induced by galactose were determined by immunoblotting with Lem8-specific antibodies. The 3-phosphoglycerate kinase (PGK) was detected as loading control.

Sequence alignment of Lem8 with four bacterial cysteine protease effectors.
The strictly conserved residues were shown in dark purple background and the Cys-His-Asp motif are marked with white letters in a red background. HopN1 and AvrpphB are from P. syringae. YopT and PfhB1 are from Y. pestis and P. multocida, respectively.
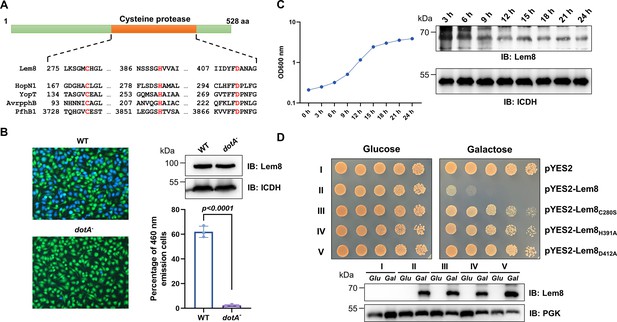
Lem8 is dispensable for intracellular growth of L. pneumophila.
(A) Subcellular distribution of Lem8. Two hours after infected with the indicated bacterial strains, bone marrow-derived macrophages (BMDMs) were immunostained using anti-Legionella antibody to identify the bacterial vacuoles (green), followed by staining with the Lem8-specific antibody (red). The nucleus was stained using Hoechst 33,342 (blue). Bar, 10 μm. (B) Intracellular growth of the Δlem8 strain in D. discoideum. D. discoideum were infected with the indicated bacterial strains at an multiplicity of infection (MOI) of 0.1, and the intracellular growth was determined at a 24-hr interval for 72 hr (left panel). The expression and translocation of Lem8 in each strain were probed with Lem8-specific antibodies (right panel). ICDH and Tubulin were used as loading controls for bacterial and host cells, respectively. Similar results were obtained in three independent experiments. (C) Intracellular growth of Δlem8 strain in BMDMs. The bacterial strains were used to infect BMDMs at an MOI of 0.1 and the intracellular growth was monitored at the indicated time points (left panel). Similar results were obtained in three independent experiments.
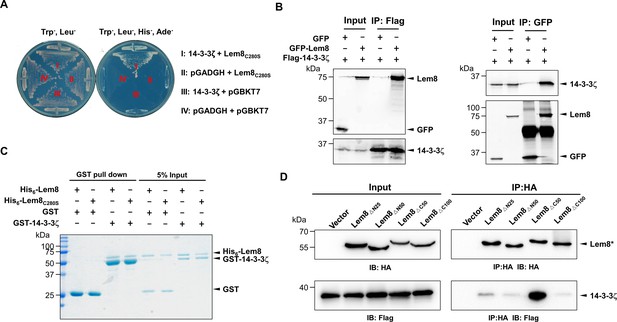
The interactions between Lem8 and 14-3-3ζ.
(A) Interactions between Lem8 and 14-3-3ζ detected by yeast two-hybrid assay. Yeast strains harboring the indicated constructs were streaked on Leu− and Trp− medium to select for plasmids (left) or on Leu−, Trp−, Ade−, and His− medium to assess the interactions (right). Images were acquired after 3-day incubation at 30°C. (B) Lem8 and 14-3-3ζ form a protein complex in mammalian cells. Total lysates of HEK293T cells transfected with the indicated plasmid combinations were immunoprecipitated with a Flag-specific antibody (left panels) or GFP-specific antibodies (right panels), and the precipitates were probed with both Flag and GFP antibodies. Similar results were obtained from at least three independent experiments and the data shown here were from one representative experiment. (C) Lem8 directly interacts with 14-3-3ζ. GST-14-3-3ζ was incubated with His6-Lem8 or His6-Lem8C280S, and the potential protein complex was captured by glutathione beads for 1 hr at 4°C. After extensive washing, bound proteins were solubilized with sodium dodecyl sulfate (SDS) loading buffer, and proteins were detected by Coomassie brilliant blue staining after being resolved by SDS/polyacrylamide gel electrophoresis PAGE. Similar results were obtained from at least three independent experiments and the data shown here were from one representative experiment. (D) Interactions between 14-3-3ζ and Lem8 deletion mutants. Lysates of 293T cells expressing Flag-14-3-3ζ and each of the HA-tagged deletion Lem8 were subjected to immunoprecipitation with the anti-HA antibody and the presence of 14-3-3ζ in the precipitates was probed with the Flag-specific antibody. Similar results were obtained from at least three independent experiments and the data shown here were from one representative experiment.
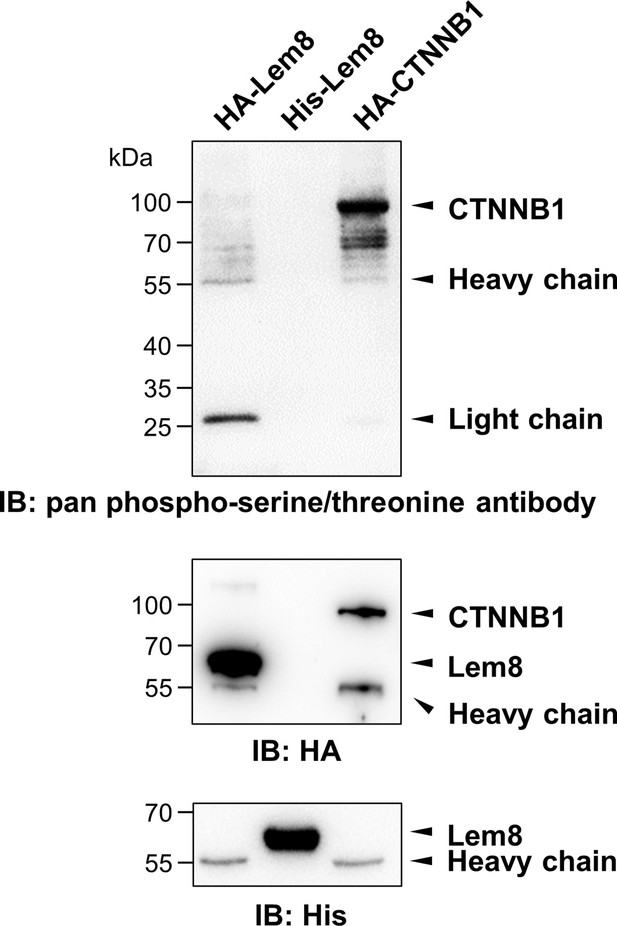
Phosphorylation of Lem8 is not required for 14-3-3ζ binding.
Lysates of HEK293T cells expressing indicated HA-tagged proteins were subjected to immunoprecipitation with agarose beads coated with the HA antibody. The precipitates, as well as His6-Lem8 purified from E. coli were resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) and probed by immunoblotting with a pan phospho-serine/threonine antibody, the HA-specific antibody and the His6-specific antibody, respectively. Results shown were one representative from three independent experiments with similar results.
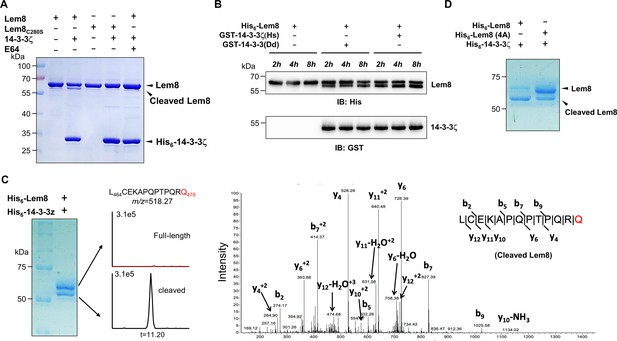
14-3-3ζ induces Lem8 to undergo self-cleavage.
(A) Self-processing of Lem8 requires 14-3-3ζ. His6-Lem8 or His6-Lem8C280S was incubated with His6-14-3-3ζ for 2 hr, proteins resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) were detected by Coomassie brilliant blue staining. The cysteine protease inhibitor E64 was added to the indicated samples at a final concentration of 10 μM. Similar results were obtained from at least three independent experiments and the data shown here were from one representative experiment. (B) The 14-3-3 protein from D. discoideum induces the self-cleavage of Lem8. His6-Lem8 was incubated with GST-14-3-3ζ or GST-14-3-3Dd for the indicated time and the mixtures separated by SDS–PAGE were detected by immunoblotting with antibodies specific for Lem8 and GST, respectively. Similar results were obtained from at least three independent experiments and the data shown here were from one representative experiment. (C) Determination of the self-cleavage site of Lem8. His6-Lem8 was incubated with His6-14-3-3ζ for 16 hr, proteins were resolved by SDS–PAGE, stained with Coomassie brilliant blue. Protein bands corresponding to full-length and cleaved Lem8 band was excised, digested with trypsin and analyzed by mass spectrometry. The detection of the semitryptic peptide -L464CEKAPQPTPQRQ476- in cleaved samples suggested that the cleavage site lies between Gln476 and Arg477. (D) Mutations in cleavage site does not abolish Lem8 self-processing. Recombinant protein of Lem8 and the 4A mutant were each incubated with His6-14-3-3ζ for 4 hr. Proteins resolved by SDS–PAGE were detected by Coomassie brilliant blue. Similar results were obtained from at least three independent experiments and the data shown here were from one representative experiment.

Identification of the self-cleavage sites of Lem8.
(A) Determination of the self-cleavage site of Lem8 by mass spectrometry. A diagram of the sequence containing the recognition site with the two diagnostic peptides used to determine the cleavage site (top panel). Protein bands corresponding to full-length and cleaved Lem8 band was excised (lower left panel), digested with trypsin and analyzed by mass spectrometry. The semitryptic peptide -L464CEKAPQPTPQRQ476- is present in cleaved samples but not in samples of full-length Lem8, whereas the fragment -A478QSLSAETER487- was only detected in samples of the full-length protein (lower right panel), supporting the notion that the cleavage site lies between Gln476 and Arg477 described in Figure 3C. (B) Self-cleavage of Lem8 removes GFP fused to its carboxyl end. GFP was fused to the indicated alleles of Lem8 and the fusion proteins were individually expressed in HEK293T cells by transfection. Samples resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) were detected by immunoblotting with GFP-specific antibodies. Results shown were one representative from three independent experiments with similar results. (C) The self-cleavage site of the 4A mutant. Protein bands from stained SDS–PAGE gels were excised and analyzed similarly as described in A. The tryptic fragment A468PQPTPAAAAQSLSAETER487- was detected only in samples prepared from the cleaved protein but not full-length protein. Identical results were obtained in multiple samples analyzed by two different mass spectrometry facilities.
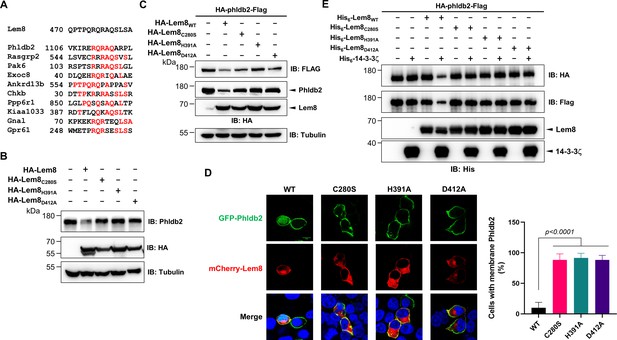
Lem8 cleaves Phldb2 in a manner that requires 14-3-3ζ.
(A) Multiple alignments of the self-cleavage site of Lem8 with potential targets in human cells identified by bioinformatic analysis. Identical residues are highlighted in red. (B) Lem8 reduces the protein levels of endogenous Phldb2 in mammalian cells. Lem8 and the indicated mutants were individually expressed in HEK293T cells by transfection. Twenty-four hours after transfection, the samples were resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) and detected by immunoblotting with anti-Phldb2 antibodies. Tubulin was used as a loading control. Results shown were one representative from three independent experiments with similar results. (C) Lem8 cleaves exogenous Phldb2 in mammalian cells. HA and Flag tag were fused to the amino and carboxyl end of Phldb2, respectively, and the double tagged protein was coexpressed in HEK293T cells with Lem8 or each of the mutants. Twenty-four hours after transfection, the samples were resolved by SDS–PAGE and probed by a HA-specific antibody and a Flag-specific antibody, respectively. Tubulin was detected as a loading control. Results shown were one representative from three independent experiments with similar results. (D) Lem8 alters the subcellular distribution of GFP fused to Phldb2. GFP was fused to the amino end of Phldb2 and the protein was coexpressed in HEK293T cells with mCherry-Lem8 or each of the mutants. Twenty-four hours after transfection, cells were fixed and nucleus were stained by Hoechst 33,342. The fluorescence Images of GFP (green), mCherry (red), and Hoechst (blue) were acquired with a Zeiss LSM 880 confocal microscope. The percentage of cells with membrane Phldb2 was calculated in Phldb2- and Lem8-positive cells (right panel). Bar, 10 μm. (E) 14-3-3ζ is required for the cleavage of Phldb2 by Lem8. HA-Phldb2-Flag was expressed in HEK293T cells, immunoprecipitated with a Flag-specific antibody, and eluted with 3× Flag peptides. Purified Phldb2 was incubated with His6-Lem8 or each of the mutants in reactions with or without His6-14-3-3ζ. Total proteins of all samples were resolved with SDS–PAGE, and probed by immunoblotting with a HA-specific antibody, a Flag-specific antibody and a His-specific antibody. Results shown were one representative from three independent experiments with similar results.
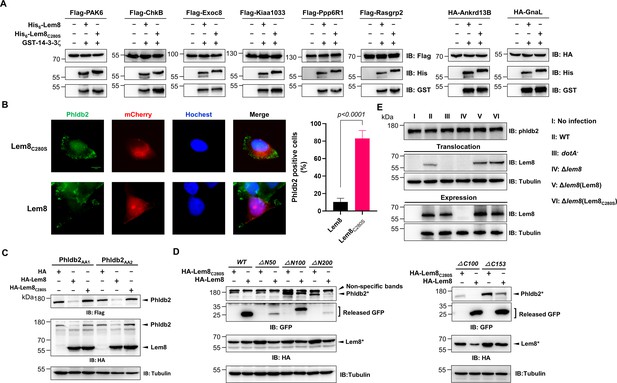
Verification of Lem8-mediated cleavage of candidate proteins and its cleavage of phldb2 at multiple sites.
(A) Cleavage of substrate candidates by Lem8. Flag- or HA-tagged Rasgrp2, Pak6, Exoc8, Ankrd13B, Chkb, Ppp6R1, Kiaa1033, Gnal, and Gpr61 expressed in HEK293T cells each was immunoprecipitated with antibodies specific for Flag or HA. Proteins eluted with 3× Flag or HA peptides were incubated with His-Lem8 and His6-14-3-3ζ. Samples resolved with sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) were probed by immunoblotting with a Flag- or HA-specific antibody. Note that Flag- or HA-GPR61 did not express so it was not examined. Results shown were one representative from three independent experiments with similar results. (B) Lem8 causes redistribution of Phldb2 in cells. Hela cells were transfected to express the indicated mCherry fusion proteins. Twenty-four hours after transfection, cells were fixed and immunostained with anti-Phldb2 antibodies. The nuclei were stained by Hoechst 33,342. Images were acquired with a Zeiss LSM 880 confocal microscope. Phldb2, green (GFP); Lem8 and its mutants, red (mCherry); nuclei, blue (Hoechst). The percentage of Phldb2-positive cells was calculated in Lem8-positive cells (right panel). Bar, 10 μm. (C) Mutations were introduced into HA-Phldb2-Flag to replace residues Arg1111 and Gln1112 (Phldb2AA1) or Gln1112 and Arg1113 (Phldb2AA2) with alanine, respectively. The two mutants were coexpressed in HEK293T cells with Lem8 or Lem8C280S. Samples were resolved with SDS–PAGE, and probed by immunoblotting with the antibody specific to HA and Flag, respectively. Mutations in the cleavage site of Phldb2 cannot completely prevent its degradation by Lem8. Results shown were one representative from three independent experiments with similar results. (D) Lem8 removes the GFP tag fused to the amino end of Phldb2 deletion mutants. GFP was fused to the amino end of Phldb2 and the indicated truncation mutants. The fusion proteins were individually coexpressed with HA-Lem8 or HA-Lem8C280S in HEK293T cells by transfection. Samples resolved by SDS–PAGE were detected by immunoblotting with GFP-specific antibodies. Results shown were one representative from three independent experiments with similar results. (E) Cleavage of Phldb2 is undetectable during L. pneumophila infection. HEK293T cells transfected to express FcγRII receptor were infected with the indicated bacterial strains. Two hours after infection, the protein levels of Phldb2, as well as the translocation and expression of Lem8, were probed with the appropriate antibodies with Tubulin and ICDH as loading control, respectively. Results shown were one representative from three independent experiments with similar results. Bacterial strains: I, noninfection; II, Lp02 (WT); III, dotA− (defective in Dot/Icm); IV, Lp02Δlem8; V, Lp02Δlem8 (pLem8); VI, Lp02Δlem8 (pLem8C280S).
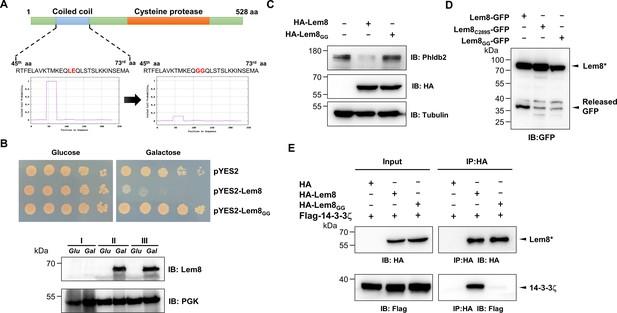
A coiled coil motif in Lem8 is important for its interactions with 14-3-3ζ.
(A) Lem8 harbors a putative coil motif. A predicted coiled coil motif located in the amino end of Lem8 (top panel). The sequence ranges from the 45th residue to the 73rd residue with a coiled coil probability of 100% according to MARCOIL (lower panel, left). Replacement of Leu58 and Glu59 with glycine (highlighted in red) is predicted to reduce the coiled coil probability to about 10% (lower panel, right). (B) The predicted coiled coil motif is critical for Lem8-mediated yeast toxicity. Yeast cells inducibly expressing Lem8 or mutant Lem8GG were serially diluted and spotted onto the indicated media for 48 hr (top panel). The expression of Lem8 and Lem8GG was examined and PGK1 was probed as a loading control (lower panel). (C) Lem8GG loses the capacity to cleave Phldb2 in mammalian cells. Lysates of HEK293T cells expressing Lem8 or Lem8GG were resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) and detected by immunoblotting with antibodies specific for Phldb2 and HA, respectively. Tubulin was used as a loading control. Results shown were one representative from three independent experiments with similar results. (D) The predicted coiled coil motif is required for self-processing of Lem8. The indicated alleles of Lem8-GFP were individually expressed in HEK293T cells by transfection. Samples resolved by SDS–PAGE were detected by immunoblotting with GFP-specific antibodies. Results shown were one representative from three independent experiments with similar results. (E) Interactions between 14-3-3ζ and the Lem8GG mutant. Lysates of 293T cells expressing Flag-14-3-3ζ with HA-Lem8 or HA-Lem8GG were subjected to immunoprecipitation with the anti-HA antibody and the presence of 14-3-3ζ in the precipitates was probed with the Flag-specific antibody. Results shown were one representative from three independent experiments with similar results.
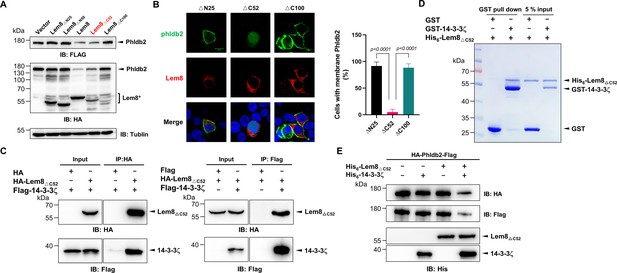
Autoprocessed Lem8 retains the cysteine protease activity.
(A) The autoprocessed form of Lem8 cleaves Phldb2 in cells. HA-Phldb2-Flag was coexpressed in HEK293T cells with Lem8 or the indicated truncation mutants including the self-processed form, Lem8∆C52. Twenty-four hours after transfection, the samples were resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) and probed by a HA-specific antibody and a Flag-specific antibody. Tubulin was used as a loading control. Results shown were one representative from three independent experiments with similar results. (B) Lem8∆C52 causes redistribution of GFP-Phldb2. Truncations of Lem8, including Lem8∆N25, Lem8∆C52, and Lem8∆C100 fused to mCherry was individually expressed in HEK293T cells with GFP-Phldb2. Twenty-four hours after transfection, the fluorescence images were acquired with a Zeiss LSM 880 confocal microscope. The percentage of cells with membrane Phldb2 was calculated in Phldb2- and Lem8-positive cells (right panel). Bar, 10 μm. (C) The interaction between Lem8∆C52 and 14-3-3ζ. Total lysates of HEK293T cells transfected with indicated plasmid combinations were immunoprecipitated with antibodies specific for HA (left panel) or Flag (right), and the precipitates were probed with both HA and Flag antibodies. Similar results were obtained from at least three independent experiments and the data shown here were from one representative experiment. (D) Lem8△C52 directly interacts with 14-3-3ζ. Mixtures containing GST-14-3-3ζ and His6-Lem8∆C52 were incubated with glutathione beads for 1 hr at 4°C. After washing, samples resolved by SDS/PAGE were detected by Coomassie brilliant blue staining. Results shown were one representative from three independent experiments with similar results. (E) The cleavage of Phldb2 by Lem8∆C52 requires 14-3-3ζ. Purified HA-Phldb2-Flag from HEK293T was incubated with His6-Lem8∆C52 in reactions with or without His6-14-3-3ζ. Total proteins of all samples were resolved with SDS–PAGE, and probed by immunoblotting with antibody specific for HA, Flag, and His6, respectively. Results shown were one representative from three independent experiments with similar results.
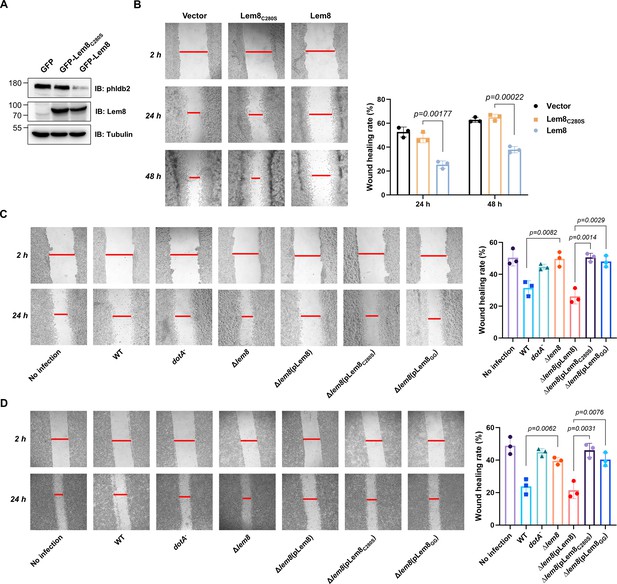
Lem8 contributes to cell migration inhibition by L. pneumophila.
(A) Establishment of cell lines stably expressing Lem8 or its enzymatically inactive mutant. HEK293T cells were transduced with lentiviral particles harboring the indicated plasmid at an multiplicity of infection (MOI) of 10 for 2 days, and the GFP-positive cells were isolated by a BD Influx cell sorter. Lysates of each cell line were probed by immunoblotting with antibodies specific for Phldb2 or Lem8. Tubulin was used as a loading control. (B) Wound-healing scratch assay of the three stable cell lines. The three cell lines were individually seeded into 6-well plates. When reached confluency, cell monolayer of each cell line was scratched using a pipette tip. Images of the wounds were captured at 2, 24, and 48 hr after making the scratches using an Olympus IX-83 fluorescence microscope. Images of a representative experiment were shown (left panel). The wound-healing rates from three independent experiments were quantitated by Image J (right panel). (C, D) Evaluation of the impact of Lem8 on cell migration in cells infected with L. pneumophila. HEK293T cells expressing the FcγII receptor or Raw264.7 cells were infected with opsonized bacteria of the indicated L. pneumophila strains at an MOI of 50 for 2 hr. After washes, the wound-healing scratch assay was performed to evaluate the impact of infection on cell migration. Images of a representative experiment were shown (C, left panel) and the wound-healing rate was analyzed by Image J (C, right panel).
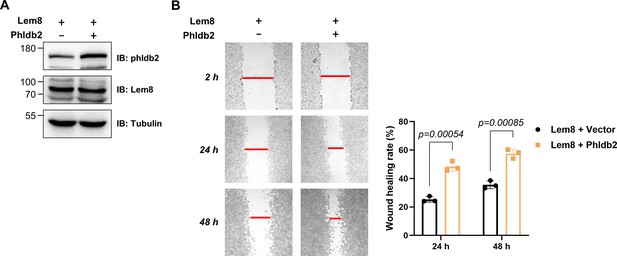
Overexpression of Phldb2 suppressed the inhibitory effects of Lem8 on cell migration.
(A) Overexpression of Phldb2 in 293T cells stably expressing Lem8. A HEK293T-derived cell line stably expressing Lem8 was transfected with empty vector or HA-Phldb2 for 18 hr, and the cell lysates were probed by immunoblotting with antibodies specific for Phldb2 or Lem8. Tubulin was used as a loading control. Results shown were one representative from three independent experiments with similar results. (B) Wound-healing scratch assay of cells expressing Phldb2. Eighteen hours after transfection, monolayers were scratched using a pipette tip. Images of the wounds were captured at 2, 24, and 48 hr, respectively. Results shown were from a representative of three independent experiments from which the quantitation of wound healing was obtained by Image J (right panel).
Additional files
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/73220/elife-73220-transrepform1-v2.docx
-
Supplementary file 1
Potential Lem8 interacting proteins identified by yeast two-hybrid screenings.
- https://cdn.elifesciences.org/articles/73220/elife-73220-supp1-v2.docx
-
Supplementary file 2
Bacterial strains, plasmids and primers used in the study.
- https://cdn.elifesciences.org/articles/73220/elife-73220-supp2-v2.docx
-
Source data 1
Source data for figures and figure supplements.
- https://cdn.elifesciences.org/articles/73220/elife-73220-data1-v2.zip