Multiple UBX proteins reduce the ubiquitin threshold of the mammalian p97-UFD1-NPL4 unfoldase
Figures
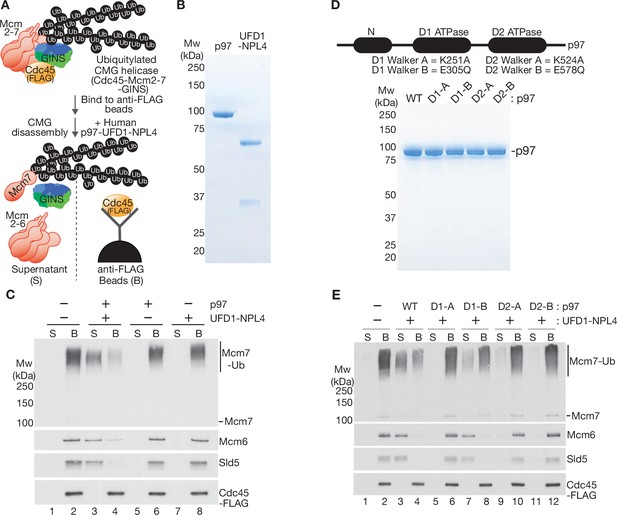
Reconstituted disassembly of ubiquitylated CMG helicase by purified human p97-UFD1-NPL4.
(A) Budding yeast CMG helicase (with an internal FLAG-tag on the Cdc45 subunit) was ubiquitylated in vitro, bound to beads coated with anti-FLAG antibody and then incubated with recombinant human p97-UFD1-NPL4 (see Materials and methods). Disassembly of ubiquitylated CMG displaced all subunits except Cdc45 into the supernatant. (B) Purified human p97 and UFD1-NPL4. (C) CMG disassembly reactions carried out according to the scheme in (A), in the presence of the indicated factors. (D) Purified recombinant human p97 with the indicated mutations in the D1 and D2 ATPase modules. (E) CMG disassembly reactions equivalent to those in (C), in the presence of the indicated factors.
-
Figure 1—source data 1
Source data for Figure 1.
- https://cdn.elifesciences.org/articles/76763/elife-76763-fig1-data1-v2.pdf
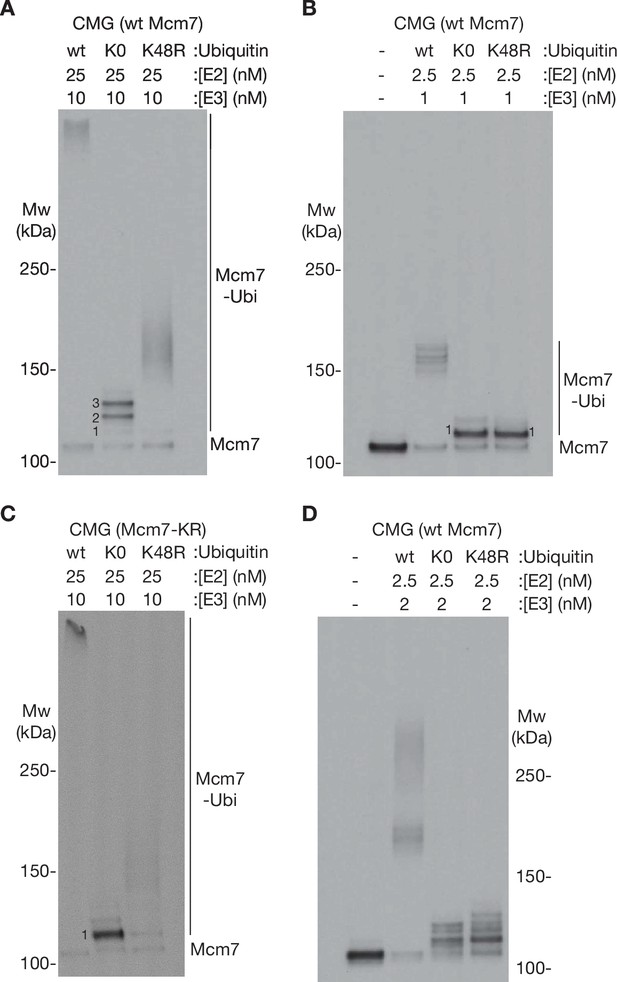
Characterisation of ubiquitin chain formation under the conditions used in this study.
(A) Reconstituted CMG ubiquitylation reactions were performed as described in Materials and methods, in the presence of 25 nM E2, 10 nM E3, and 6 µM of the indicated form of ubiquitin (wt [wild type], K0 = ubiquitin with all lysines mutated to arginine, K48R = ubiquitin with K48R mutation). (B) Similar reactions were performed in the presence of 2.5 nM E2 and 1 nM E3. (C) CMG ubiquitylation reactions were performed as in (A) but with CMG containing an allele of Mcm7 (Mcm7-KR, described in Materials and methods) with surface lysine mutations that restrict ubiquitylation to a single chain per CMG complex. (D) Reactions performed as in (A, B) but with 2.5 nM E2 and 2 nM E3.
-
Figure 1—figure supplement 1—source data 1
Source data for Figure 1—figure supplement 1.
- https://cdn.elifesciences.org/articles/76763/elife-76763-fig1-figsupp1-data1-v2.pdf
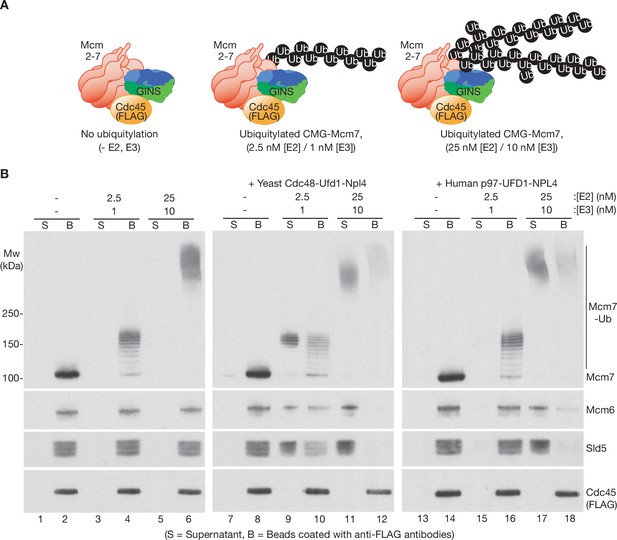
Human p97-UFD1-NPL4 has a much higher ubiquitin threshold than yeast Cdc48-Ufd1-Npl4.
(A) Reconstituted CMG ubiquitylation reactions were performed under conditions that produced a single chain of up to ~12 ubiquitins on CMG-Mcm7 (2.5 nM E2, 1 nM E3; see Figure 1—figure supplement 1B), or 2–3 chains per CMG-Mcm7 (25 nM E2, 10 nM E3; see Figure 1—figure supplement 1A). (B) The products of the ubiquitylation reactions in (A) were bound to beads coated with anti-FLAG antibodies, before incubating as indicated with yeast Cdc48-Ufd1-Npl4 or human p97-UFD1-NPL4. CMG disassembly was monitored via displacement of subunits from beads (B) to supernatant (S), except for FLAG-tagged Cdc45 that remained bound to the anti-FLAG beads.
-
Figure 2—source data 1
Source data for Figure 2.
- https://cdn.elifesciences.org/articles/76763/elife-76763-fig2-data1-v2.pdf
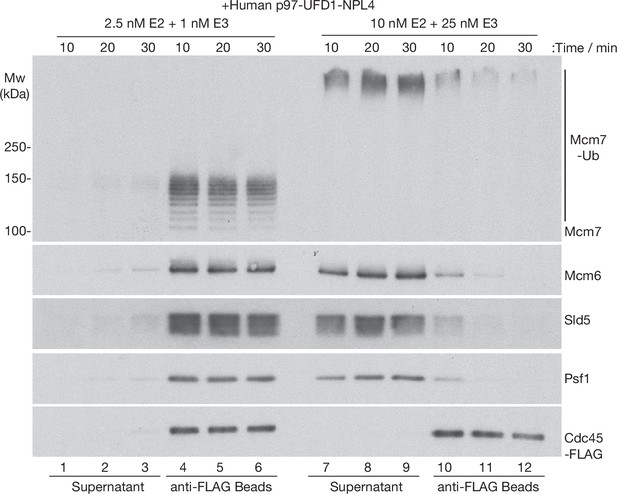
Time course analysis of CMG disassembly reactions by human p97-UFD1-NPL4.
CMG ubiquitylation reactions were performed as in Figure 2, with the indicated concentrations of E2 and E3 enzymes. Subsequently, ubiquitylated CMG was bound to anti-FLAG beads before incubation with human p97-UFD1-NPL4 for the indicated times. Disassembly of ubiquitylated CMG was reflected by release of CMG subunits into the supernatant, except for Cdc45-FLAG that remained bound to the anti-FLAG beads.
-
Figure 2—figure supplement 1—source data 1
Source data for Figure 2—figure supplement 1.
- https://cdn.elifesciences.org/articles/76763/elife-76763-fig2-figsupp1-data1-v2.pdf
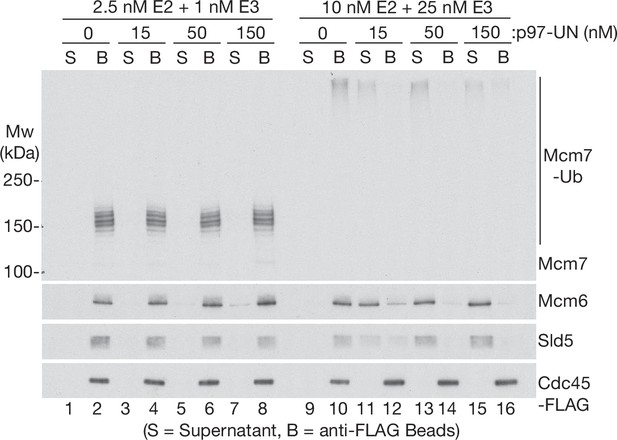
Human p97-UFD1-NPL4 preferentially disassembles extensively ubiquitylated CMG helicase.
CMG ubiquitylation and disassembly reactions were performed as above, except that the disassembly reactions contained the indicated concentrations of p97-UFD1-NPL4, in addition to 15 nM ubiquitylated CMG substrate. Disassembly of ubiquitylated CMG was reflected by release of helicase subunits into the supernatant, apart from Cdc45-FLAG that remained bound to the anti-FLAG beads.
-
Figure 2—figure supplement 2—source data 1
Source data for Figure 2—figure supplement 2.
- https://cdn.elifesciences.org/articles/76763/elife-76763-fig2-figsupp2-data1-v2.pdf
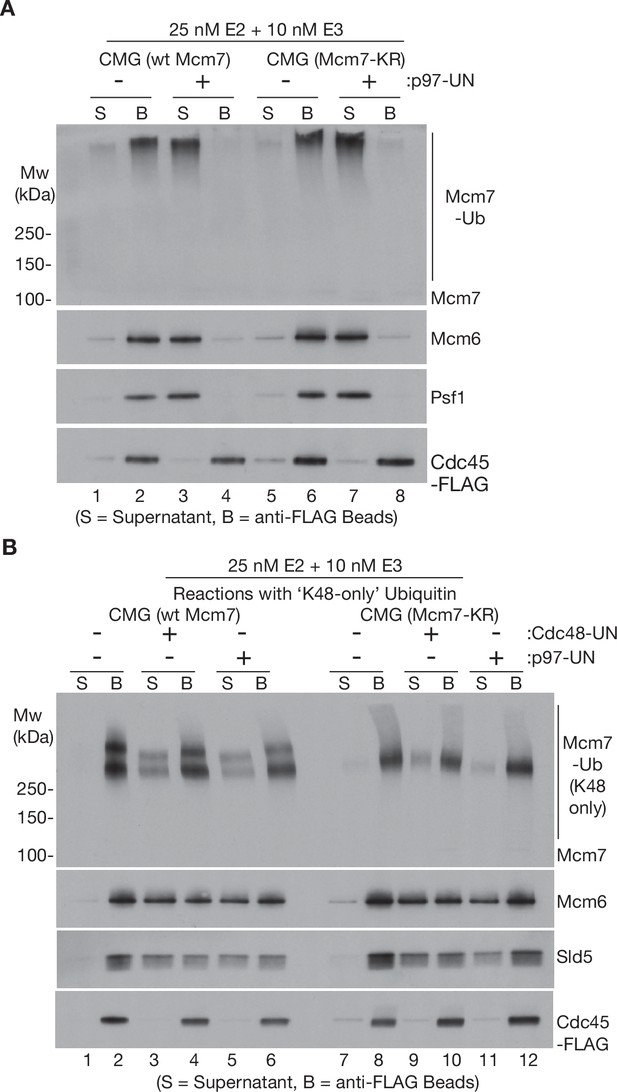
Disassembly of CMG helicase by human p97-UFD1-NPL4 does not require multiple ubiquitin chains, or other ubiquitin chain linkages apart from K48.
(A) Wild-type CMG and CMG-Mcm7-KR were ubiquitylated as in Figure 1 A + C and then bound to beads coated with anti-FLAG antibody. Disassembly reactions with then carried out as described in Figure 1, in the presence or absence of human p97-UFD1-NPL4 as indicated. (B) CMG (wt Mcm7) and CMG (Mcm7-KR) were ubiquitylated as above, except that the reactions contained ‘K48-only’ ubiquitin with all lysines mutated to arginine except for lysine 48. Disassembly reactions were then carried out as above, in the presence of yeast Cdc48-Ufd1-Npl4 or human p97-UFD1-NPL4 as indicated. Note that a fraction of the unfolded Mcm7 subunits with long ‘K48-only’ ubiquitin chains associated non-specifically with the beads in this experiment, so disassembly was most easily monitored via displacement into the supernatant of other CMG subunits such as Mcm6 and Sld5.
-
Figure 2—figure supplement 3—source data 1
Source data for Figure 2—figure supplement 3.
- https://cdn.elifesciences.org/articles/76763/elife-76763-fig2-figsupp3-data1-v2.pdf
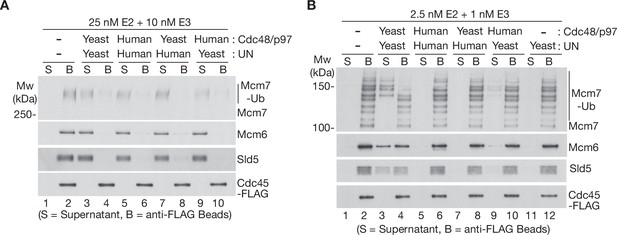
The Ufd1-Npl4/UFD1-NPL4 adaptor complex determines the ubiquitin threshold of Cdc48/p97.
(A) CMG helicase was ubiquitylated as in Figure 1—figure supplement 1A, then bound to anti-FLAG beads. Disassembly reactions were then performed as indicated in the presence of yeast Cdc48 or human p97, combined with yeast Ufd1-Npl4 or human UFD1-NPL4. (B) Analogous reactions with CMG ubiquitylated as in Figure 1—figure supplement 1B.
-
Figure 3—source data 1
Source data for Figure 3.
- https://cdn.elifesciences.org/articles/76763/elife-76763-fig3-data1-v2.pdf

Sequence alignment of human and yeast orthologues of p97 and UFD1-NPL4.
(A) Human p97 and yeast Cdc48 were aligned with Clustal Omega software and the output viewed with MView (Madeira et al., 2019). (B, C) Similar analysis for NPL4/Npl4 and UFD1/Ufd1.
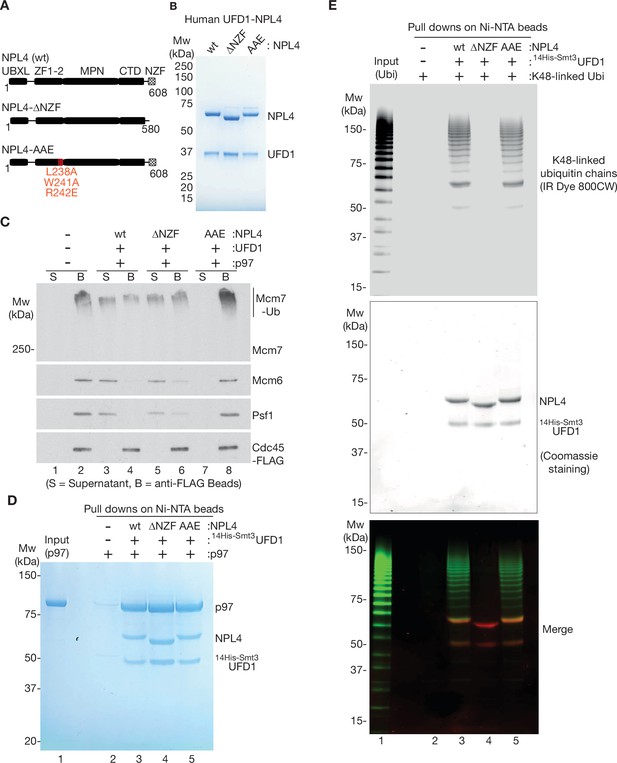
The NPL4 groove is essential for disassembly of ubiquitylated CMG by p97-UFD1-NPL4, whereas the NZF domain is largely dispensable.
(A) Domain analysis of human NPL4 UBXL = UBX like domain that interacts with a p97 N-terminal domain; ZF1-2 are Zn fingers that anchor the MPN domain to p97; MPN is the ‘Mpr1/Pad1 N-terminal domain’ that sits on top of the D1 ring of p97 and contains the NPL4 groove that in yeast Npl4 binds an unfolded ubiquitin intermediate; CTD = C-Terminal Domain that interacts with folded ubiquitin; NZF = NPL4 ubiquitin-binding Zn finger. The NPL4-∆NZF allele lacks amino acids 581–608. The NPL4-AAE allele has three mutations at conserved sites in the NPL4 groove (L238A, W241A, and R242E), which in yeast Npl4 contact an unfolded ubiquitin intermediate and are essential for the initiation of substrate unfolding (Twomey et al., 2019). (B) Human UFD1 and indicated versions of NPL4 were expressed as recombinant proteins in E. coli, before purification of UFD1-NPL4 complexes (see Materials and methods). (C) CMG was ubiquitylated as in Figure 1—figure supplement 1A, before binding to anti-FLAG beads. Disassembly reactions were performed as above, in the presence of the indicated factors. (D) Purified human p97 was mixed with variants of UFD1-NPL4 as indicated, before binding to Ni-NTA beads via His-tagged UFD1 (see Materials and methods). (E) Purified K48-linked ubiquitin chains were conjugated to the fluorescent label ‘IR Dye 800CW’ as described in Materials and methods. The labelled chains were then mixed with the indicated variants of UFD1-NPL4, before binding to Ni-NTA beads via His-tagged UFD1 (Materials and methods provides details). Bound ubiquitin chains were detected by fluorescence (top panel) and bound UFD1-NPL4 was detected by staining with Coomassie blue.
-
Figure 3—figure supplement 2—source data 1
Source data for Figure 3—figure supplement 2.
- https://cdn.elifesciences.org/articles/76763/elife-76763-fig3-figsupp2-data1-v2.pdf
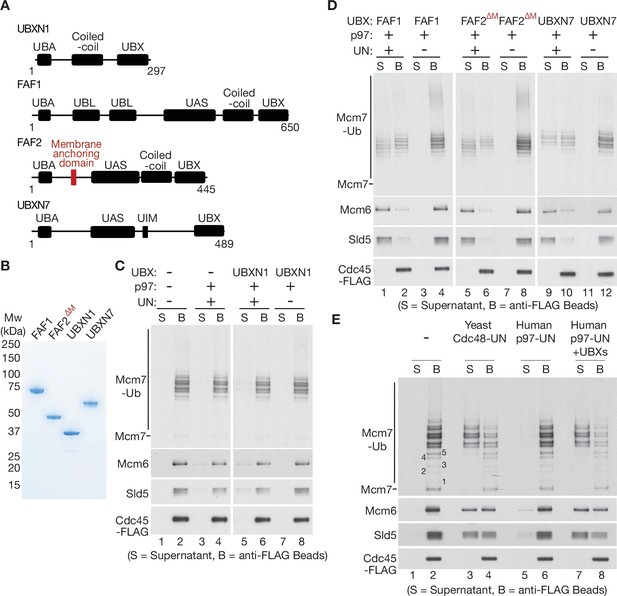
UBXN7, FAF1, and FAF2 reduce the ubiquitin threshold of p97-UFD1-NPL4.
(A) Domain organisation of the indicated UBX proteins. UBA = ‘UBiquitin-Associated’ domain that binds ubiquitin; UBX = ‘UBiquitin regulatory X’ domain that binds p97; UBL = ‘UBiquitin-Like’ domain that binds ubiquitin and NEDD8; UAS = domain of unknown function in FAF1/FAF2/UBXN7. (B) Purified proteins – the membrane anchoring domain of FAF2 was deleted in FAF2∆M to facilitate expression of a soluble protein. (C–E) CMG disassembly reactions in the presence of the indicated factors, performed as described in Figures 1—3. See also Figure 4—figure supplements 1–3.
-
Figure 4—source data 1
Source data for Figure 4.
- https://cdn.elifesciences.org/articles/76763/elife-76763-fig4-data1-v2.pdf
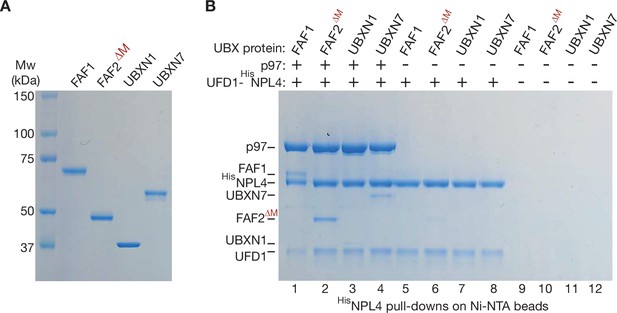
Purified human UBX proteins bind to p97-UFD1-NPL4.
(A) Purified UBX proteins. (B) Proteins were mixed as indicated and the factors associated with 6HisNPL4 were isolated on Ni-NTA beads as described in Materials and methods. The bound proteins were monitored by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE) and Coomassie staining.
-
Figure 4—figure supplement 1—source data 1
Source data for Figure 4—figure supplement 1.
- https://cdn.elifesciences.org/articles/76763/elife-76763-fig4-figsupp1-data1-v2.pdf
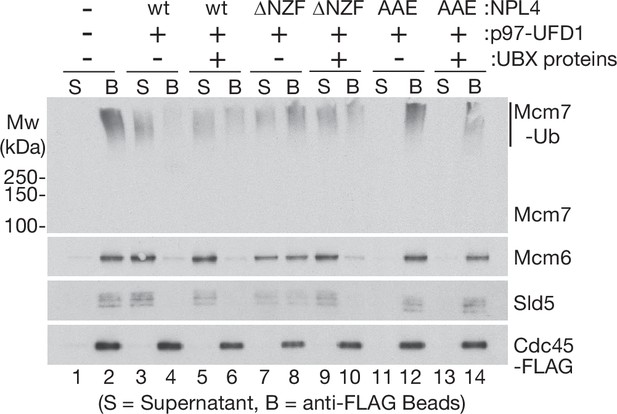
Stimulation of p97-UFD1-NPL4 activity by UBX proteins requires the NPL4 groove and suppresses loss of the ubiquitin-binding NPL4-NZF.
CMG ubiquitylation is performed as in Figure 1—figure supplement 1A. Disassembly reactions were then carried out as in Figure 1, in the presence of the indicated factors. Variants of NPL4 correspond to those in Figure 3—figure supplement 2A, B.
-
Figure 4—figure supplement 2—source data 1
Source data for Figure 4—figure supplement 2.
- https://cdn.elifesciences.org/articles/76763/elife-76763-fig4-figsupp2-data1-v2.pdf
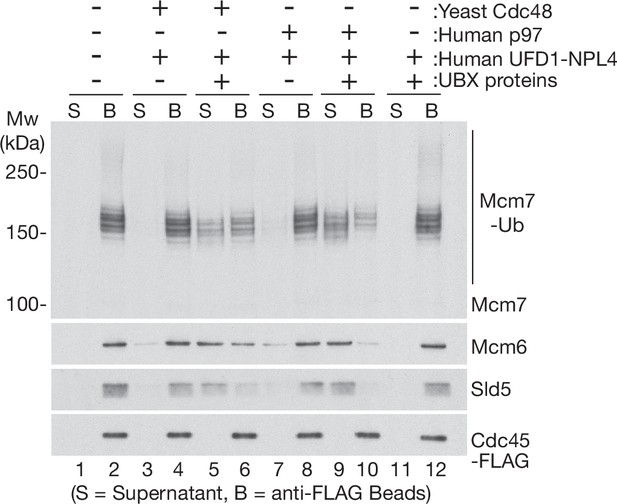
Human UBX proteins stimulate CMG helicase disassembly by yeast Cdc48 in the presence of human UFD1-NPL4.
CMG was ubiquitylated as in Figure 1—figure supplement 1D, before binding to anti-FLAG beads. Disassembly reactions were then performed in the presence of the indicated factors.
-
Figure 4—figure supplement 3—source data 1
Source data for Figure 4—figure supplement 3.
- https://cdn.elifesciences.org/articles/76763/elife-76763-fig4-figsupp3-data1-v2.pdf
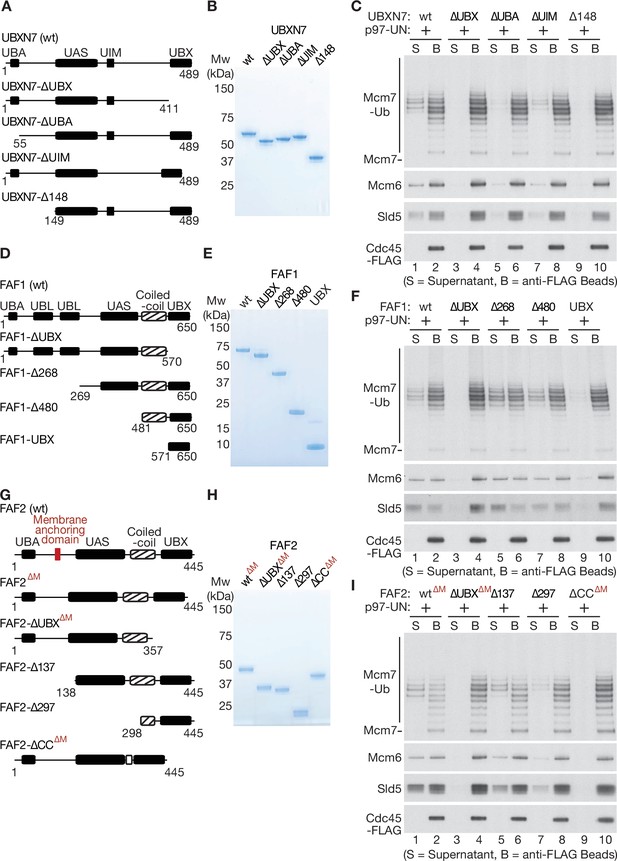
Mapping domains of UBXN7, FAF1, and FAF2 that stimulate the unfoldase activity of p97-UFD1-NPL4.
(A) Truncations of UBXN7. (B) Purified proteins. (C) CMG was ubiquitylated as in Figure 1—figure supplement 1D and then bound to anti-FLAG beads. Disassembly reactions were performed as above, in the presence of the indicated factors. (D–F) Equivalent analysis for FAF1. (G–I) Analogous truncations of FAF2 – ‘∆M’ indicates alleles that contain the amino terminus of the protein but lack the membrane anchoring domain. For (A), (D), and (G), the numbers correspond to residues in the full-length proteins. Domains were predicted using the SMART algorithm (http://smart.embl-heidelberg.de/) and Alphafold (Jumper et al., 2021).
-
Figure 5—source data 1
Source data for Figure 5.
- https://cdn.elifesciences.org/articles/76763/elife-76763-fig5-data1-v2.pdf
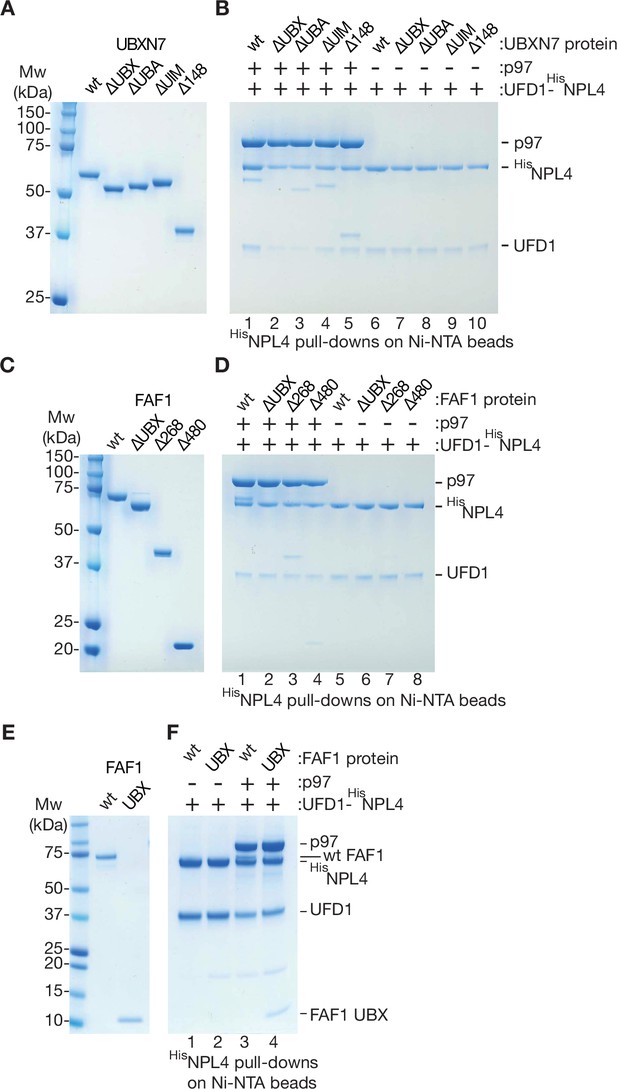
Association of UBXN7 and FAF1 truncations with p97-UFD1-NPL4.
(A) Purified truncated versions of UBXN7, as in Figure 5A. (B) Association of UBXN7 truncations with p97-UFD1-NPL4 was monitored by pull downs of 6HisNPL4 on Ni-NTA beads, as in Figure 4—figure supplement 1 (C–F) Analogous analysis for the indicated truncations of FAF1.
-
Figure 5—figure supplement 1—source data 1
Source data for Figure 5—figure supplement 1.
- https://cdn.elifesciences.org/articles/76763/elife-76763-fig5-figsupp1-data1-v2.pdf
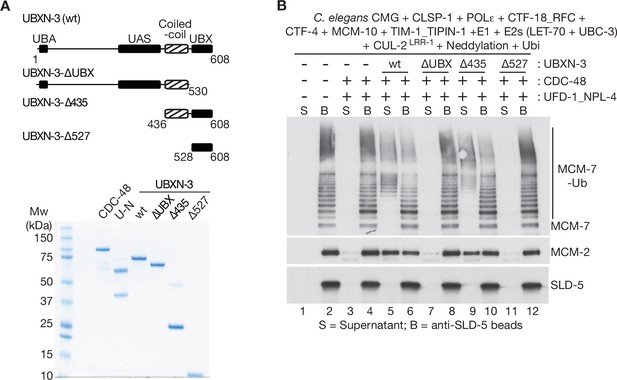
The coiled-coil and UBX domains of C.elegans UBXN-3 are required to stimulate the unfoldase activity of CDC-48_UFD-1_NPL-4.
(A) The indicated truncations of C. elegans UBXN-3 (upper panel) were purified together with C. elegans CDC-48 and UFD-1_NPL-4 (lower panel; UN = UFD-1_NPL-4). (B) C. elegans CMG was ubiquitylated in the presence of the indicated factors and then bound to beads coated with antibodies to the SLD-5 subunit of GINS, as described in Materials and methods. Ubiquitylated CMG was then incubated as shown with C. elegans CDC-48, UFD-1_NPL-4 and wt or truncated forms of UBXN-3. CMG disassembly was monitored by release of ubiquitylated MCM-7 (MCM-7-Ub) and MCM-2 into the supernatant.
-
Figure 5—figure supplement 2—source data 1
Source data for Figure 5—figure supplement 2.
- https://cdn.elifesciences.org/articles/76763/elife-76763-fig5-figsupp2-data1-v2.pdf
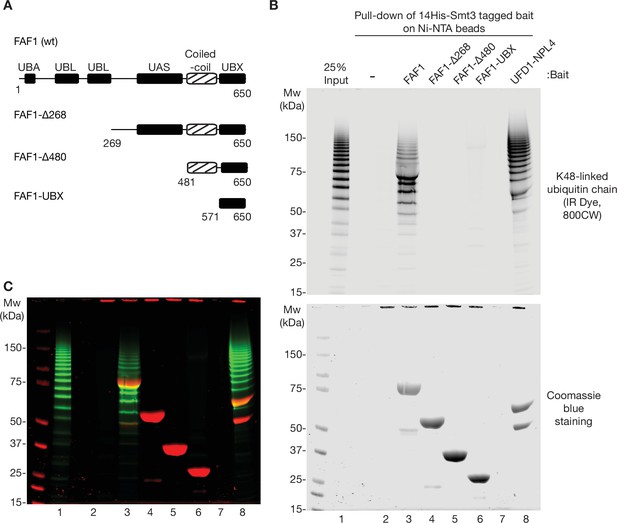
Ubiquitin binding of human FAF1.
(A) Domain structure of human FAF1 and associated truncations, as in Figure 5D. (B) As in Figure 3—figure supplement 2, the indicated His-tagged bait proteins (14His-Smt3FAF1 or 14His-Smt3UFD1-NPL4) were pre-bound to Ni-NTA beads, then incubated with K48-linked ubiquitin chains that were labelled with the fluorescent dye IR Dye 800CW. Bound ubiquitin chains were monitored by fluorescence (upper panel) whilst the bait proteins were detected via Coomassie blue staining (lower panel). (C) Merge of images from (B).
-
Figure 5—figure supplement 3—source data 1
Source data for Figure 5—figure supplement 3.
- https://cdn.elifesciences.org/articles/76763/elife-76763-fig5-figsupp3-data1-v2.pdf
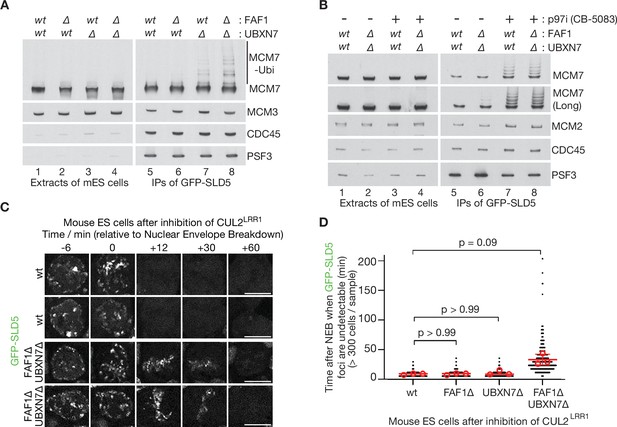
Partial redundancy between UBXN7 and FAF1 for CMG helicase disassembly during S-phase and mitosis in mouse ES cells.
(A) CMG was isolated from extracts of mouse embryonic stem cells (mES) with the indicated genotypes, by immunoprecipitation of GFP-tagged SLD5 subunit of the helicase. (B) Equivalent experiment in which cells were treated as indicated with 5 µM CB-5083 (p97i = p97 inhibitor) for 3 hr before harvesting. (C) Time-lapse video analysis of mitotic entry in mouse ES cells expressing GFP-SLD5, following inhibition of CUL2LRR1. Scale bars correspond to 10 µm. (D) Quantification of the data in (C). The individual data points from three independent experiments (>300 cells in total) are depicted as black dots in a scatter plot. The mean values from each experiment are shown in red circles, whilst red bars and error bars represent the average of the mean values and the associated standard deviations (n = 3). The samples were then compared by a Kruskal–Wallis test followed by Dunn’s test, yielding the indicated p values. See also Figure 6—figure supplements 3–5 (note that Figure 6—figure supplement 4E confirms that the difference between FAF1∆ UBXN7∆ and cells lacking either FAF1 or UBXN7 is statistically significant, despite the weaker significance of the data in (D)).
-
Figure 6—source data 1
Source data for Figure 6.
- https://cdn.elifesciences.org/articles/76763/elife-76763-fig6-data1-v2.pdf

Sequence alignment of human and mouse orthologues of p97, NPL4, and UFD1.
(A) Human and mouse orthologues of p97 were aligned with Clustal Omega software and the output viewed with MView (Madeira et al., 2019). (B, C) Similar analysis for NPL4 and UFD1.
Sequence alignment of human and mouse orthologues of FAF1, FAF2, and UBXN7.
(A) Human and mouse orthologues of FAF1 were aligned with Clustal Omega software and the output viewed with MView (Madeira et al., 2019). (B, C) Similar analysis for FAF2 and UBXN7.
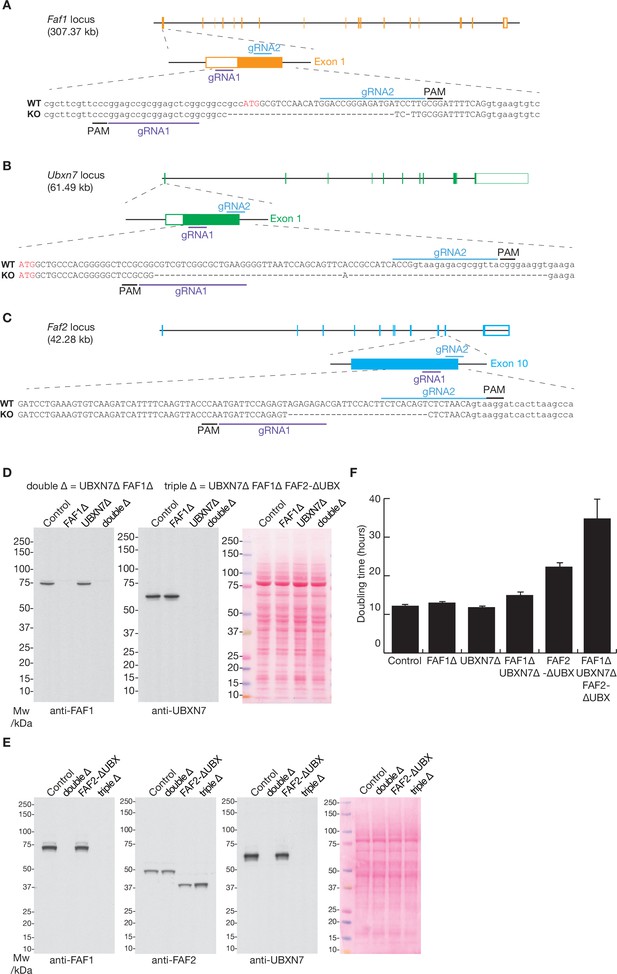
The deletion of the Faf1, Ubxn7, and Faf2 genes by CRISPR-Cas9.
(A–C) Loci and corresponding guide RNAs (gRNAs) that were used to target the D10A ‘nickase’ mutant of Cas9 to the Faf1, Ubxn7, and Faf2 loci in mouse ES cells. See Materials and methods for further details. PAM = ‘Protospacer Adjacent Motif’ that takes the form ‘NGG’ and is essential for cutting by Cas9. (D) Immunoblot analysis with the indicated antibodies to monitor single and double deletion of the Faf1 and Ubxn7 genes. The right panel shows total protein stained with Ponceau S. (E) Analogous immunoblot analysis of the indicated strains, including deletion of the UBX domain of FAF2. (F) Doubling time of mES cells with the indicated genotypes was monitored as described in Materials and methods. The histogram indicates the mean values from three independent experiments, together with the associated standard deviations.
-
Figure 6—figure supplement 3—source data 1
Source data for Figure 6—figure supplement 3.
- https://cdn.elifesciences.org/articles/76763/elife-76763-fig6-figsupp3-data1-v2.pdf
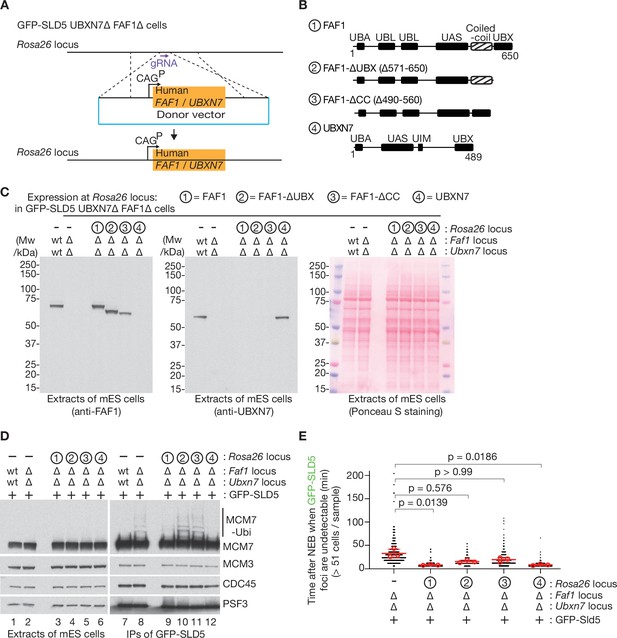
Rescue of FAF1∆ UBXN7∆ mouse ES cells by expression of human FAF1 or UBXN7.
(A) Scheme for expressing human FAF1 or UBXN7 from the CAG promoter at the Rosa26 locus in mouse ES cells. A donor vector was integrated as shown, in GFP-SLD5 UBXN7∆ FAF1∆ cells. (B) Three truncations of FAF1 were chosen for expression, together with full-length UBXN7. (C) Immunoblot analysis (left and centre panels) of extracts of cells with the indicated genotypes. The right panel provides a loading control via Ponceau S staining of the membrane after protein transfer. (D) As in Figure 6A, B, CMG was isolated from extracts of mouse ES cells with the indicated genotypes, by immunoprecipitation of the GFP-tagged SLD5 subunit of the helicase. The indicated proteins were monitored by immunoblotting. (E) Quantification of time-lapse video analysis of mitotic entry in mouse ES cells expressing GFP-SLD5, following inhibition of CUL2LRR1, equivalent to Figure 6C, D. As discussed in Figure 6D, the individual data for three separate experiments (>320 cells in total per genotype) are plotted as black dots in a scatter plot, together with mean values (red circles), average of means (red bars), and associated standard deviation (error bars, n = 3). The samples were compared by a Kruskal–Wallis test followed by a Dunn’s test, yielding the indicated p values. Note that the data for FAF1∆ UBXN7∆ control cells, lacking a construct at Rosa26 to express FAF1 or UBXN7, are taken from Figure 6D.
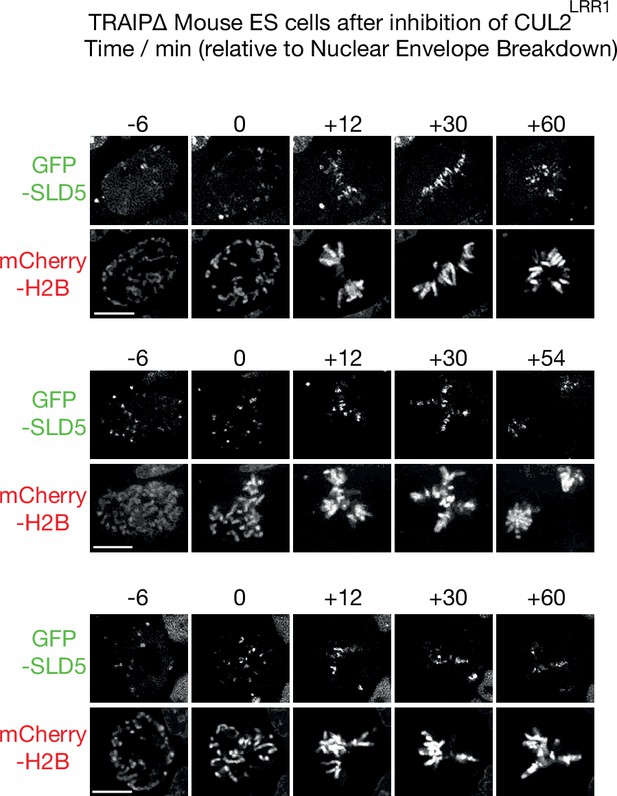
Mitotic CMG disassembly is blocked in mES cells that lack TRAIP.
TRAIP∆ cells expressing GFP-SLD5 from the endogenous Gins4 (Sld5) locus were grown as in Figure 6C and monitored by time-lapse video analysis. Three examples are shown of mitotic entry following inhibition of CUL2LRR1, illustrating the persistence of GFP-SLD5 on mitotic chromatin throughout the observed time course (100% of cells, n = >20 cells per sample). Scale bars correspond to 10 µm.
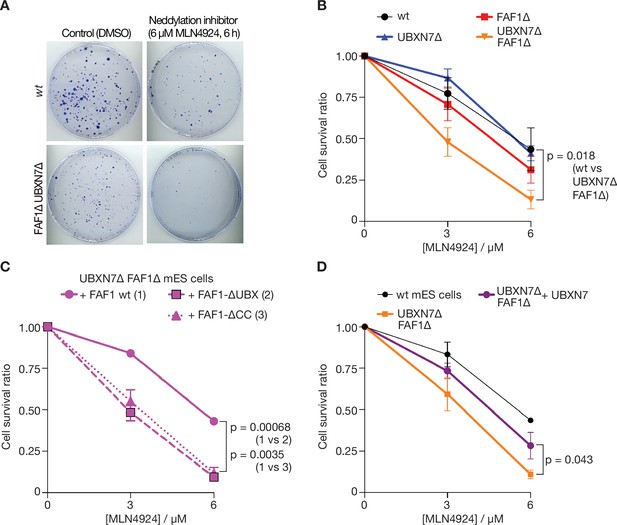
Cells lacking FAF1 and UBXN7 are sensitive to global inhibition of cullin ligase activity.
(A) Cells of the indicated genotypes were treated as shown and then grown on 10 cm plates for 6 days. Surviving colonies were fixed and stained with crystal violet. (B) Viability of cells treated with 0, 3, or 6 µM MLN-4924 for 6 hr. The data represent the mean values and standard deviation for three independent experiments (n = 3). The mean survival ratios of wt and FAF1∆ UBXN7∆ after treatment with 6 µM MLN-4924 condition were compared via a two-tailed t-test. (C) Analogous experiment involving FAF1∆ UBXN7∆ mouse ES cells expressing the indicated versions of human FAF1 (as in Figure 6—figure supplement 4B) at the Rosa26 locus. (D) Similar experiment to that in (C), comparing wt mouse ES cells, FAF1∆ UBXN7∆ cells, and FAF1∆ UBXN7∆ cells expressing human UBXN7 from the Rosa26 locus.
Additional files
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/76763/elife-76763-transrepform1-v2.pdf
-
Supplementary file 1
Reagents and resources used in this study.
- https://cdn.elifesciences.org/articles/76763/elife-76763-supp1-v2.docx
-
Supplementary file 2
Plasmids generated in this study.
- https://cdn.elifesciences.org/articles/76763/elife-76763-supp2-v2.docx





