A framework for community curation of interspecies interactions literature
Figures

Increase of molecular pathogen-host interaction publications and gene-phenotype information during the last 35 years curated in the Pathogen–Host Interactions database (PHI-base).
Gray bars show the number of publications in the Web of Science Core Collection database retrieved with search terms ‘(fung* or yeast) and (gene or factor) and (pathogenicity or virulen* or avirulence gene*).’ Black vertical bars show the number of articles retrieved from PubMed (searching on title and abstract). White and black triangles show the number of curated plant and animal pathogen genes, respectively.
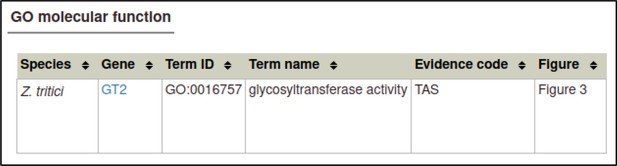
Schematic representation of pathogen–host interactions.
(a) The disease triangle illustrates the requirement for the correct abiotic and biotic environmental conditions to ensure disease when an adapted pathogen encounters a suitable host. (b) A non-gene-for-gene genetic relationship where compatible interactions result in disease on all host genotypes (depicted as genotypes 1–4), but the extent of disease formation is influenced to a greater or lesser extent by the presence or absence of a single pathogen virulence gene product X. In host genotypes 1 and 3, the pathogen gene product X is the least required for disease formation. The size of each black oval in each of the eight genetic interactions indicates the severity of the disease phenotype observed, with a larger oval indicating greater severity. (c) A gene-for-gene genetic relationship. In this genetic system, considerable specificity is observed, which is based on the direct or indirect interaction of a pathogen avirulence (Avr) effector gene product with a host resistance (R) gene product to determine specific recognition (an incompatible interaction), which is typically observed in biotrophic interactions (Jones and Dangl, 2006). In one scenario, the product of the Avr effector gene binds to the product of the R gene (a receptor) to activate host resistance mechanisms. In another scenario, the product of the Avr effector gene binds to an essential host target which is guarded by the product of the R gene (a receptor). Once Avr effector binding is detected, host resistance mechanisms are activated. The absence of the Avr effector product or the absence of the R gene product leads to susceptibility (a compatible interaction). The small black dot indicates no disease formation, and the large black oval indicates full disease formation. (d) An inverse gene-for-gene genetic relationship. Again, considerable specificity is observed based on the interaction of a pathogen necrotrophic effector (NE) with a host susceptibility (S) target to determine specific recognition. The product of the pathogen NE gene binds to the product of the S gene (a receptor) to activate host susceptibility mechanisms.

Conceptual model showing the relationship between metagenotypes, genotypes, and annotations.
The curator selects a pathogen genotype and a host genotype to combine into a metagenotype. The metagenotype can be annotated with pathogen–host interaction phenotypes from PHIPO (the Pathogen–Host Interaction Phenotype Ontology).

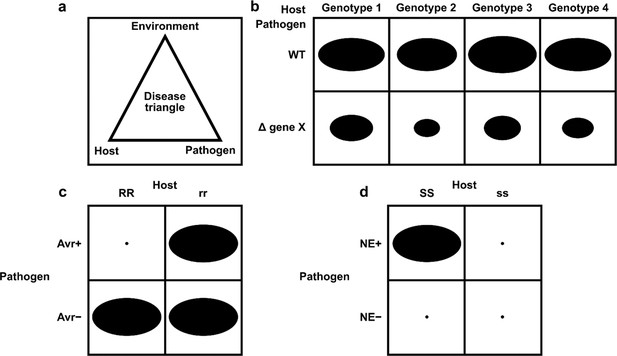
Canto entity-relationship model.
Simplified UML class diagram showing the relations between entities (things of interest) in a Canto curation session. The numbers on the connecting lines represent the cardinality of the relation, meaning how many of one entity can be related to another entity: 0...n means ‘zero or more;’ 1...n means ‘one or more.’ Lines with a hollow arrowhead indicate that the target entity (at the head of the arrow) is a generalization of the source entity (at the tail of the arrow). Boxes outlined in bold indicate new entities which were added to support curation in the Pathogen–Host Interaction Community Annotation Tool (PHI-Canto).

Entity–relationship model for the main Canto database.
This database stores data that is shared across all curation sessions. Database tables are represented as boxes, and arrows between boxes indicate a connection between tables. The table and property names contain numerous abbreviations, which are expanded as follows: curs: curation session, pub: publication, db: database, xref: cross-reference, cv: controlled vocabulary.
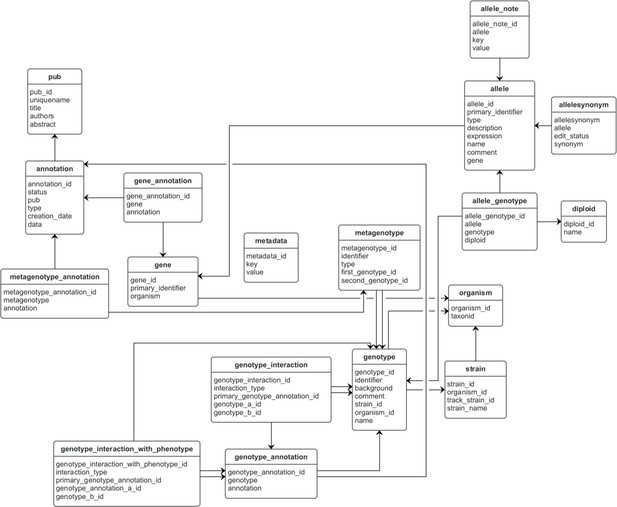
Entity–relationship model for a Canto curation session database.
This database stores data that is unique to a curation session. Database tables are represented as boxes, and arrows between boxes indicate a connection between tables. The ‘pub’ table stands for ‘publication.’
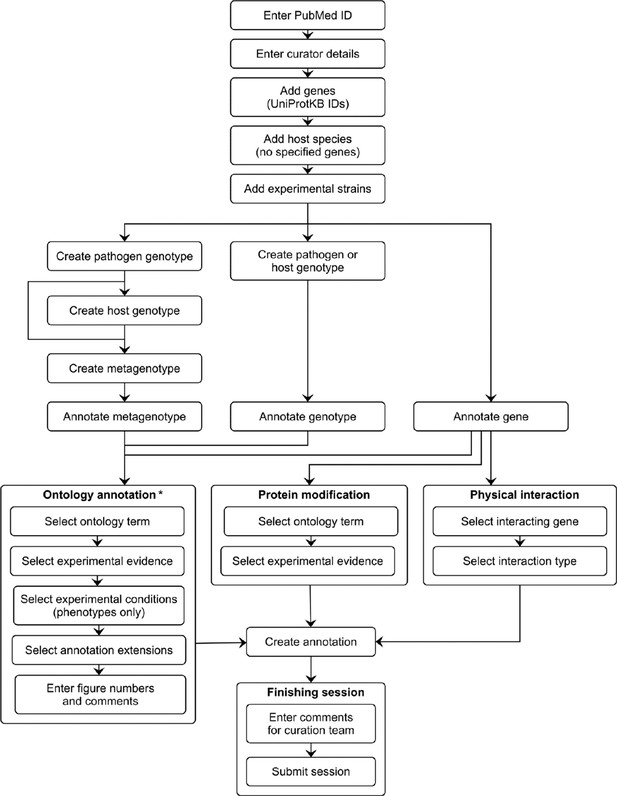
Pathogen–Host Interaction Community Annotation Tool (PHI-Canto) curation workflow diagram.
This diagram shows the curation workflow from the start of a curation session to its submission. The PubMed ID of the publication to be curated is entered and the title is automatically retrieved. The curator enters their name, email address, and ORCID iD. On the species and genes page, the experimental pathogen and host genes are entered using UniProtKB accession numbers, and for experiments where a mutant pathogen genotype is assayed on a wild-type host with no specified genes, there is the option to select the host species from an autocomplete menu. Information on the specific experimental strains used for each species is entered. After entering this initial information, the curator follows one of three distinct workflows depending on the biological feature the user wants to annotate (metagenotype, genotype, or gene annotation type). Except for genes, biological features are created by composing less complex features: genotypes from alleles (generated in the pathogen or host genotype management pages), and metagenotypes from genotypes (generated in the metagenotype management page). Biological features are annotated with terms from a controlled vocabulary (usually an ontology), plus additional information that varies based on the annotation type. The curator has the option to generate further annotations after creating one, but this iterative process is not represented in the diagram for the sake of brevity. After all annotations have been made, the session is submitted into the Pathogen–Host Interactions database (PHI-base) version 5. * Note that the 'Ontology annotation' group covers multiple annotation types, all of which annotate biological features with terms from an ontology or controlled vocabulary. These annotation types are described in Table 1.
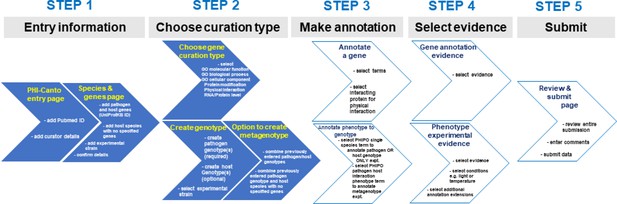
Alternative curation step workflow.
The flow diagram represents the Pathogen–Host Interaction Community Annotation Tool (PHI-Canto) curation process from beginning to end in five steps. This diagram is an alternative representation to the image depicted in Figure 4. During step 2 of the workflow, the curator chooses either the gene annotation or genotype/metagenotype annotation process. Multiple annotations can be made using both annotation processes which can then be submitted for review.

What you need to curate a publication using the Pathogen–Host Interaction Community Annotation Tool (PHI-Canto).
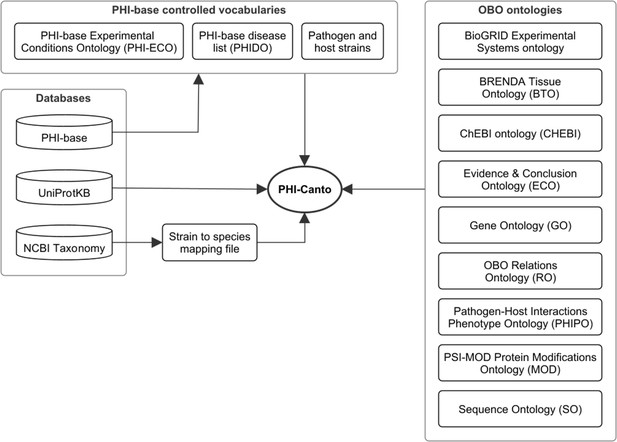
Network diagram showing the data resources used by the Pathogen–Host Interaction Community Annotation Tool (PHI-Canto).
Of the databases shown, the Pathogen–Host Interactions database (PHI-base) provides data (experimental conditions, disease names, and species strain names) used to create terms in the PHI-base controlled vocabularies; the UniProt Knowledgebase (UniProtKB) provides accession numbers for proteins that PHI-Canto uses to identify genes; and the NCBI Taxonomy database is used to generate a mapping file relating taxonomic identifiers lower than species rank to their nearest taxonomic identifiers at species rank. The OBO ontologies group contains ontologies in the OBO format that PHI-Canto uses for its annotation types. The parenthesized text after the ontology name indicates the term prefix for the ontology.

Resources relied upon by the Pathogen–Host Interaction Community Annotation Tool (PHI-Canto).

The interspecies curation framework and the interoperability of the Pathogen–Host Interaction Community Annotation Tool (PHI-Canto).
(a) The interspecies curation framework consists of three main components. First, a curation tool called PHI-Canto, second, a new species-neutral phenotype ontology called PHIPO (the Pathogen–Host Interaction Phenotype Ontology), and thirdly, a selection of additional controlled vocabularies for disease names (PHIDO), experimental conditions (PHI-ECO), pathogen and host species, and natural strains associated with each species. The two-way arrows indicate that terms from the ontology and controlled vocabularies are used in curation with PHI-Canto, and that new terms required for curation may be suggested for inclusion within the ontology and controlled vocabularies. (b) The PHI-Canto and PHIPO content curation framework (gray box) uses persistent identifiers and cross-referenced information from UniProt, Ensembl Genomes, and the Gene Ontology. PHIPO is made available at the OBO Foundry. Newly minted wild-type gene annotations are suggested for inclusion into the Gene Ontology via the EBI Gene Ontology Annotation database. Data curated in PHI-Canto, following expert review, is then shared with ELIXIR data resources such as UniProtKB, Ensembl Genomes, FungiDB, and KnetMiner, and provided on request to other databases (FgMutantDB, GloBI). Researchers can look up curated information via the Pathogen–Host Interactions database (PHI-base) web interface or can download the whole dataset from PHI-base for inclusion in their bioinformatics pipelines. Authors can submit data to PHI-base by curating their publications into PHI-Canto. The origin of data is indicated by directional arrows.
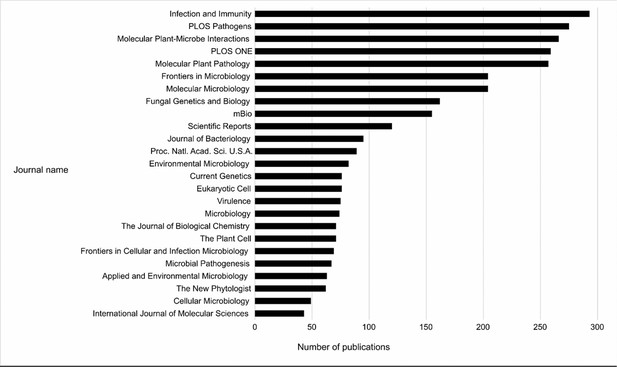
Top 25 Journals in the Pathogen–Host Interactions database (PHI-base).
Bar chart showing the top 25 journals by number of publications curated in PHI-base, as of version 4.13 (published May 9, 2022). Publication counts were generated by extracting every unique PubMed identifier (PMID) from PHI-base, then using the Entrez Programming Utilities (E-Utilities) to retrieve the journal name for each PMID, and finally summing the count of journal names. The total number of journals in version 4.13 of PHI-base was 291.
Pathogen-host interaction phenotype for ‘unaffected pathogenicity’.
Note: Phenotype annotations use evidence codes modeled on the Evidence & Conclusion Ontology (ECO).
Evidence code ‘Cell growth assay’ corresponds to ‘cell growth assay evidence’ (ECO:0001563).

Pathogen-host interaction phenotype for ‘altered pathogenicity or virulence’.
Note: Phenotype annotations use evidence codes modeled on the Evidence & Conclusion Ontology (ECO).
Evidence code ‘Macroscopic observation (qualitative observation)’ corresponds to the new ECO term ‘qualitative macroscopy evidence’ (ECO:0006342).
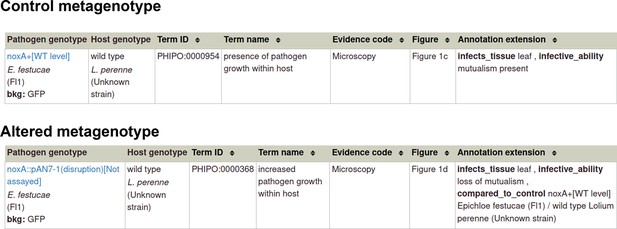
Pathogen-host interaction phenotype: Example 1 Illustrating a phenotype associated with the pathogen component within the Pathogen-Host Interaction.
Note: Phenotype annotations use evidence codes modeled on the Evidence & Conclusion Ontology (ECO). Evidence code ‘Microscopy’ corresponds to ‘microscopy evidence’ (ECO:0001098).

Pathogen-host interaction phenotype: Example 2 Illustrating a phenotype associated with the host component within the Pathogen-Host Interaction.

Gene Ontology (GO) biological process annotation for ‘a pathogen effector’.
Note: ‘Effector-mediated suppression of host pattern-triggered immunity’ (GO:0052034) is a descendant term of ‘effector-mediated modulation of host process by symbiont’ (GO:0140418).
Note: GO annotations use GO evidence codes (http://geneontology.org/docs/guide-go-evidence-codes/).
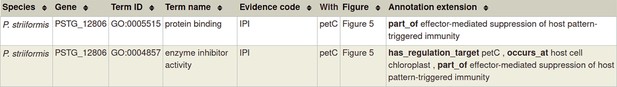
Gene Ontology (GO) molecular function annotation for ‘a pathogen effector’.
Please note that in the case of a physical interaction (protein–protein interaction) between the pathogen and host gene products (PSTG_12806 and PetC in the example above, respectively) this information can be curated using the Physical Interaction curation workflow, documented in https://canto.phi-base.org/docs/physical_interaction_annotation.
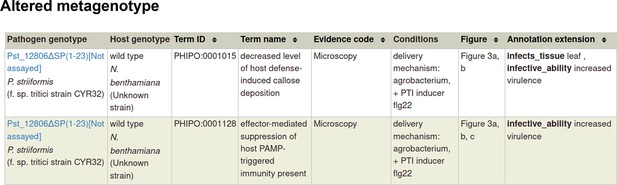
Pathogen-host interaction phenotypes for ‘a pathogen effector’.
In this case, there are no metagenotype control annotations. This is because it is not possible to create and annotate a metagenotype comprising of an empty vector control within the pathogen component of the metagenotype.

Gene Ontology (GO) biological process annotation for ‘a pathogen effector’ within ‘a gene-for-gene interaction’.

Gene Ontology (GO) molecular function annotation for ‘a pathogen effector’ within ‘a gene-for-gene interaction’.

Gene Ontology (GO) biological process annotation for ‘a pathogen necrotrophic effector’ within ‘an inverse gene-for-gene interaction’.

Gene Ontology (GO) molecular function annotation for ‘a pathogen necrotrophic effector’ within ‘an inverse gene-for-gene interaction’.

Gene-for-gene phenotype annotations for ‘an inverse gene-for-gene interaction’.
Note: The Annotation extensions (AEs) capture the detail of what has occurred within the pathogen-host interactions.

Pathogen phenotype.
Please note that in this curation example, no AEs were required.

The Pathogen–Host Interaction Community Annotation Tool (PHI-Canto) homepage provides a text field where publications can be entered by providing their PubMed ID (PMID).
The PMID in this case is 29020037.

The Pathogen–Host Interaction Community Annotation Tool (PHI-Canto) will automatically retrieve details of the publication from PubMed so that the curator can confirm that they have entered the correct PubMed ID (PMID).
After accepting the publication, the curator is prompted for their name, email address, and (optionally) an ORCID ID, which are used to attribute the curation to the curator, and to contact the curator in case of problems with the curation session.
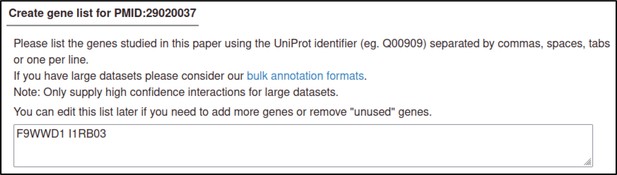
The gene is the most basic unit of annotation in the Pathogen–Host Interaction Community Annotation Tool (PHI-Canto): every other biological feature that can be annotated involves a gene, so genes are entered first.
PHI-Canto uses accession numbers from the UniProt Knowledgebase (UniProtKB) to uniquely identify proteins for the genes of interest in the curated publication. The UniProtKB accession numbers for the publication are shown.

Since this publication describes a wild-type host species (T. aestivum) with no specified genes of interest, the curator must add the host to the session by entering its NCBI Taxonomy ID in a separate field.

The Pathogen–Host Interaction Community Annotation Tool (PHI-Canto) automatically retrieves details of the proteins from UniProtKB, including the gene name, gene product, and taxonomy (e.g. the species name).

The curator must enter the strains for each organism studied in the publication or must specify when the strain was not known (or not specified in the publication).
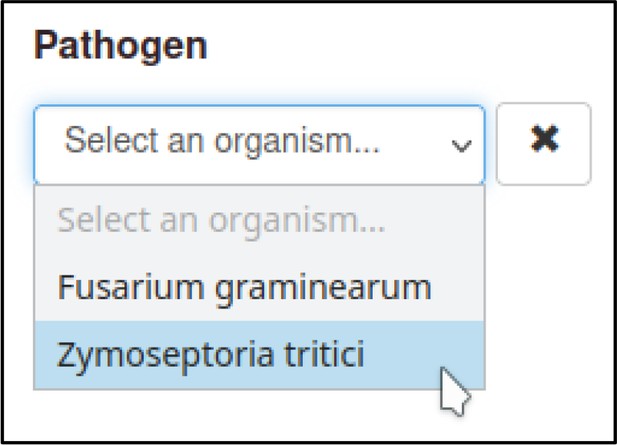
The Pathogen–Host Interaction Community Annotation Tool (PHI-Canto) provides a pre-populated list of strains for many species that the curator can select from, though they also have the option to specify a strain not in the list as free text. In this publication, the pathogen strains are PH-1 for F. graminearum and IPO323 for Z. tritici. Two cultivars of T. aestivum were used: cv. Bobwhite and cv. Riband.

In order to show that deleting GT2 in the pathogen causes a loss of pathogenicity, the curator must annotate the interaction between the mutant pathogen and its host with a phenotype, meaning the interaction must be added to the curation session.
In the Pathogen–Host Interaction Community Annotation Tool (PHI-Canto), interactions are represented as metagenotypes, which are the combined genotypes of the pathogen and host species. Before the curator can create a metagenotype, they must first create a genotype. Genotypes are composed of alleles (except in the case of wild-type host genotypes with no specified genes, as described later), and metagenotypes are composed from genotypes. So, the curator must first create an allele from a gene, then a genotype from an allele, then a metagenotype from two genotypes. The curator starts from the Pathogen genotype management page, following a link from the Curation summary page.
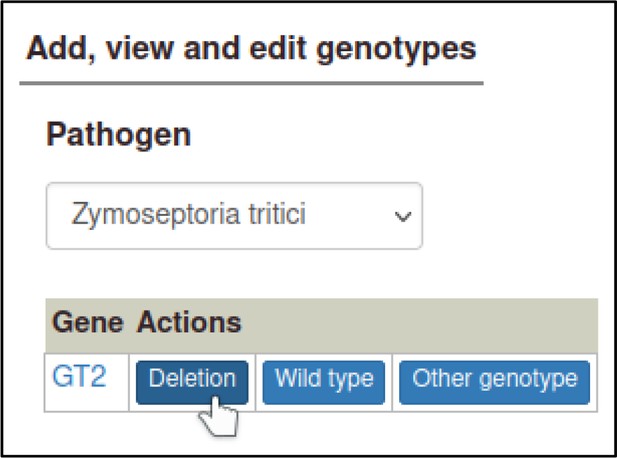
Selecting a pathogen species shows a list of genes for the species, with buttons to create types of alleles.
Here, the curator selects ‘Deletion’ for a deletion allele.
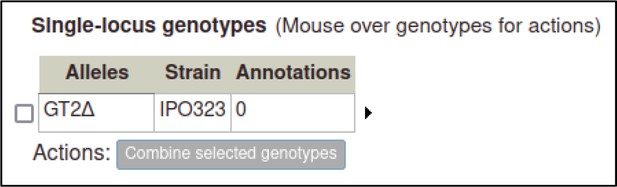
After selecting this, the Pathogen–Host Interaction Community Annotation Tool (PHI-Canto) creates a genotype containing a single allele, with the allele name automatically generated from the gene name followed by a delta symbol.
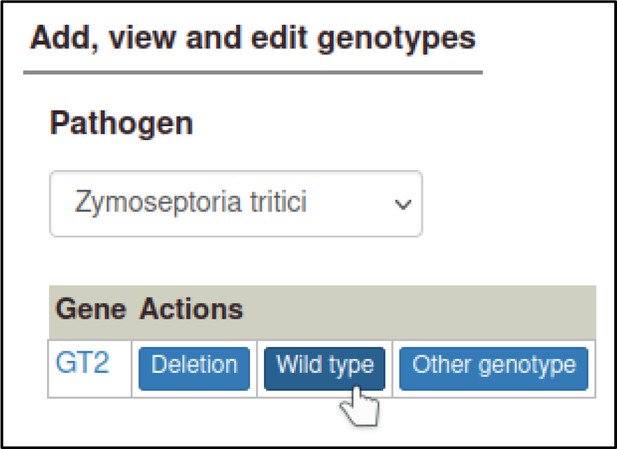
The curator will also need to prepare a wild-type genotype for the pathogen GT2 gene, which can be added to the control metagenotype so that any changes in the phenotype (between the wild-type pathogen and the altered pathogen inoculated onto the host) can be properly annotated.
This first requires making a wild-type allele for GT2, using the ‘Wild-type’ allele type.

Wild-type alleles require the gene expression level to be specified.
In this case, there was no change in expression level, so the curator selects ‘Wild-type product level.’ The Pathogen–Host Interaction Community Annotation Tool (PHI-Canto) automatically creates an allele name by appending a plus symbol to the gene name.
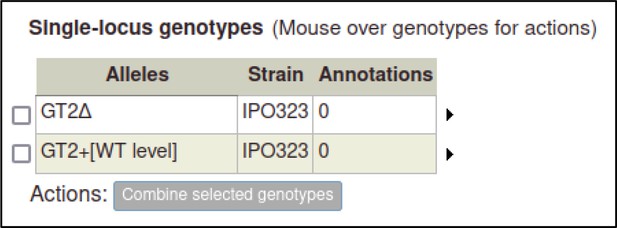
As genotypes are created, they are added to a table of genotypes on their respective genotype management page (Pathogen genotype management for pathogens, Host genotype management for hosts).
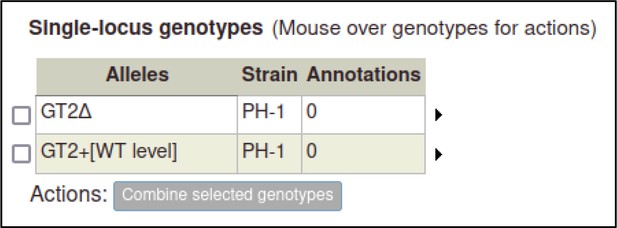
The curator can repeat the process above to create pathogen genotypes for F. graminearum.

Metagenotypes are created using the Metagenotype management page, where genotypes previously added to the curation session can be combined into a metagenotype.
The curator can reach this page from the Curation Summary page, or from either the pathogen, or host genotype management page.
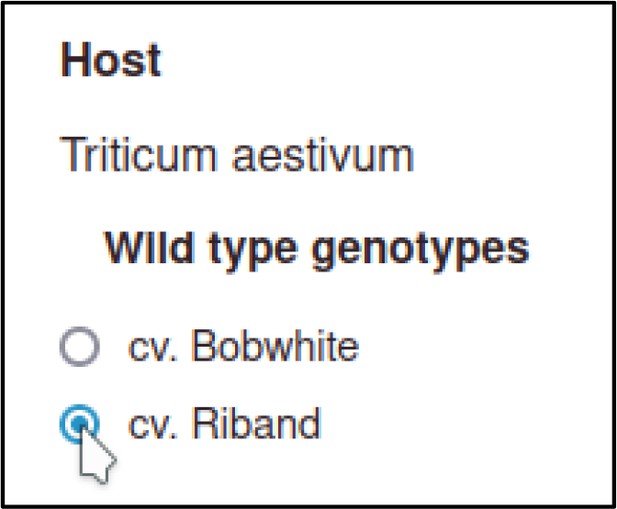
Then the curator selects a host genotype.
For wild-type hosts, the Pathogen–Host Interaction Community Annotation Tool (PHI-Canto) provides a shortcut where a strain can be selected without needing to create an allele as part of the genotype.

The curator selects ‘Make metagenotype’ to create the metagenotype for the interaction.
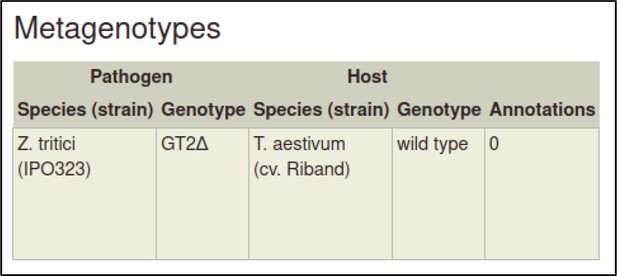
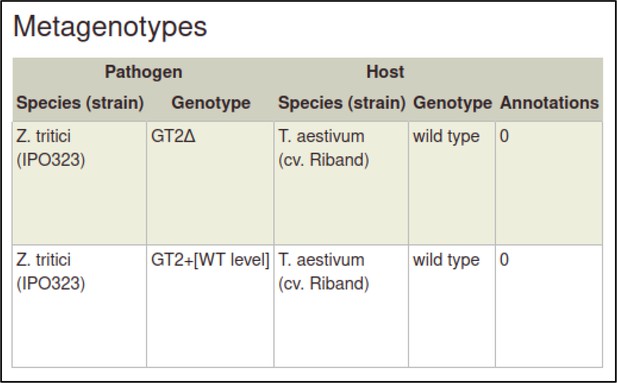
The metagenotype is displayed in a table as a combination of pathogen and host genotype.
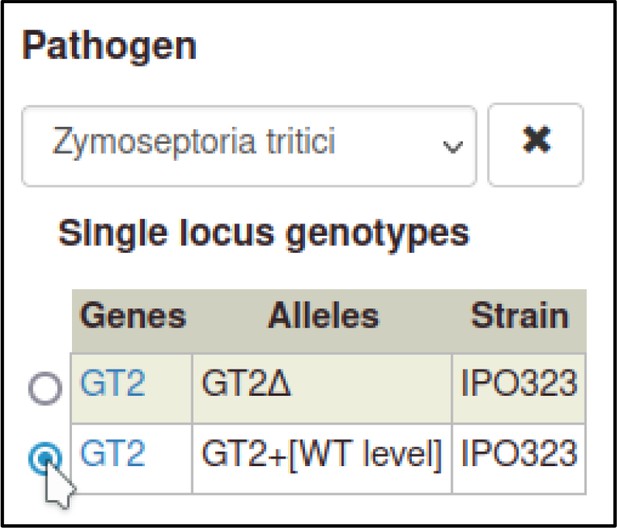
This process can be repeated to create the metagenotype for the wild-type interaction between Z. tritici and T. aestivum.
In this case, the pathogen genotype containing the wild-type GT2 is selected instead of the deletion allele.

Creating the corresponding metagenotypes for F. graminearum and T. aestivum simply requires changing the pathogen species and selecting cv. Bobwhite for the host strain.
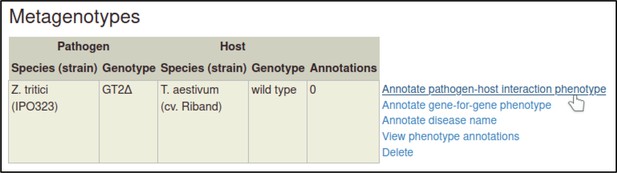
Metagenotypes can be annotated with phenotypes by selecting the ‘Annotate pathogen-host interaction phenotype’ action.
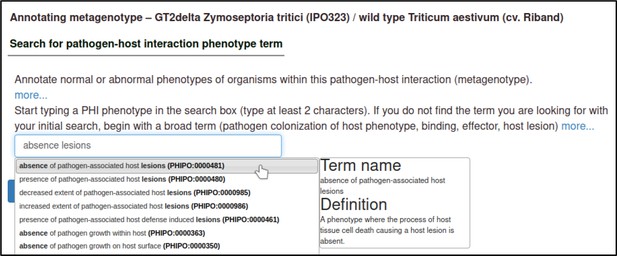
The first step is to select a term from a controlled vocabulary that describes the phenotype of the interaction.
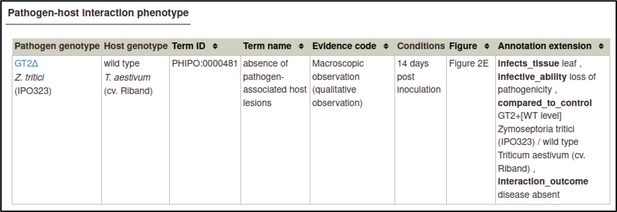
The Pathogen–Host Interaction Community Annotation Tool (PHI-Canto) uses terms from the Pathogen–Host Interaction Phenotype Ontology (PHIPO) for this purpose. The primary observed phenotype, in this case, is the absence of pathogen-associated host lesions (PHIPO:0000481).
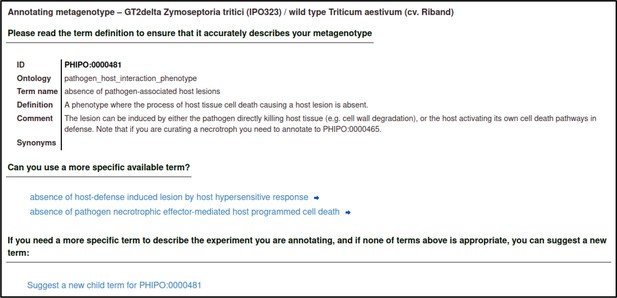
Upon selecting the term, the curator is shown a description of the term and its synonyms to help confirm that their chosen term is appropriate.

The curator must select an evidence code for the observation of the phenotype.
In this case, the phenotype was observed macroscopically, and measured qualitatively.
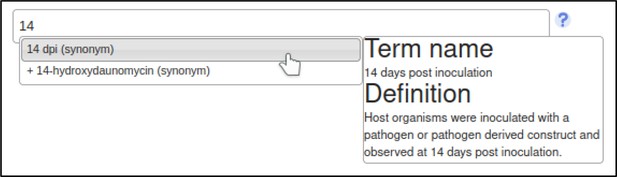
The curator may also specify experimental conditions for the experiment – such as the growth medium, or days elapsed after inoculation of the host.
This annotation specifies that the assay was performed 14 days after inoculation with the Z. tritici GT2 deletion mutant.

The Pathogen–Host Interaction Community Annotation Tool (PHI-Canto) uses annotation extensions to provide additional information about the conditions and outcome of the pathogen–host interaction.
Of particular note are the host tissue infected, the changes to the infective ability of the pathogen, the presence (or absence) of disease, and the interaction used as a control for the interaction involving a mutant pathogen.
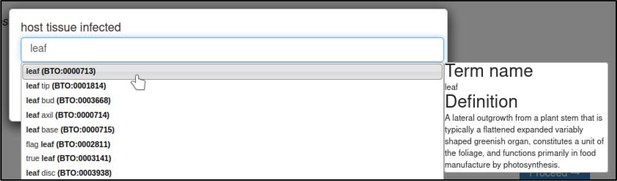
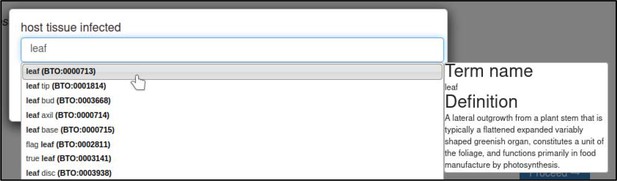
The host tissue that was infected during the interaction is annotated with the ‘host tissue infected’ annotation extension.
This extension uses ontology terms from the BRENDA Tissue Ontology (BTO). In this case, the curator specifies that the leaf (BTO:0000713) of T. aestivum was infected.
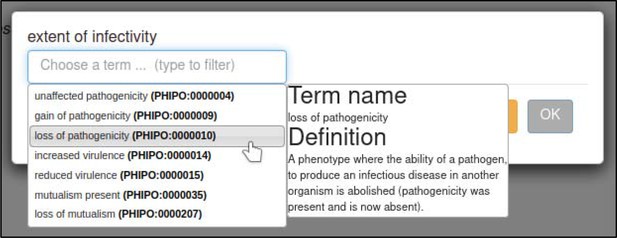
Changes in the infective ability of the pathogen are annotated with the ‘extent of infectivity’ annotation extension.
This extension uses a subset of ontology terms from the Pathogen–Host Interaction Phenotype Ontology (PHIPO). In this case, the curator specifies that the interaction resulted in a loss of pathogenicity (PHIPO:0000010).
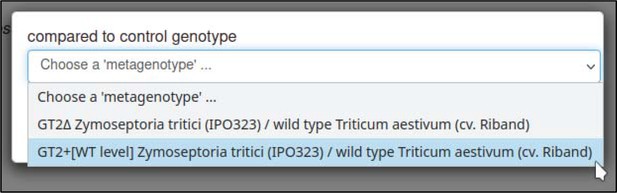
The control interaction (to which the interaction being annotated should be compared) can be annotated with the ‘compared to control genotype’ annotation extension.
This annotation allows any metagenotype in the curation session to be designated as a control. In this case, the curator selects the wild-type metagenotype that was created earlier.

The presence or absence of disease resulting from the interaction can be annotated with the ‘outcome of interaction’ annotation extension.
This extension uses a subset of ontology terms from the Pathogen–Host Interaction Phenotype Ontology (PHIPO). In this case, the curator specifies that no disease was observed as a result of the interaction: disease absent (PHIPO:0001199).

After adding annotation extensions, the curator has the option to provide the figure number from the publication (if any) that illustrates the phenotype.
In this case, the figure was Figure 2E.
The curator can also provide additional information in a comments field, in case of details that are not appropriate for any other field.
Once the above steps are completed, the phenotype annotation is created.

The above annotation can be used as a template for the interaction between the wild-type pathogen and host, since many of the variables are the same.
The Pathogen–Host Interaction Community Annotation Tool (PHI-Canto) provides a ‘Copy and edit’ feature that allows curators to use one annotation as a template for creating another.
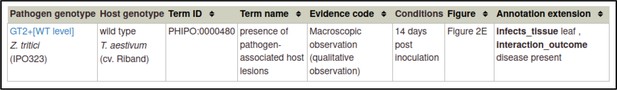
For the wild-type interaction, the pathogen genotype is changed to wild-type GT2, the phenotype term is changed to presence of pathogen-associated host lesions (PHIPO:0000480), the interaction outcome is changed to disease present (PHIPO:0001200), and the extensions for infective ability and control metagenotypes are removed, since they are not applicable.

The interaction between Z. tritici and T. aestivum can also be used as a template for the interaction between F. graminearum and T. aestivum.
Here, the pathogen genotype is changed to the GT2 deletion F. graminearum, the host strain is changed to cv. Bobwhite, the experimental condition is changed to ‘13 days post inoculation,’ the host tissue infected is changed to inflorescence (BTO:0000628), the control metagenotype is updated accordingly, and the figure number is changed to 4E.

The changes required for the wild-type interaction between F. graminearum and T. aestivum are the same as those required for Z. tritici and T. aestivum, since the interaction outcome is the same (presence of pathogen-associated host lesions, and presence of disease).

Shown here is a table of all the pathogen–host interaction phenotypes from this curation example.
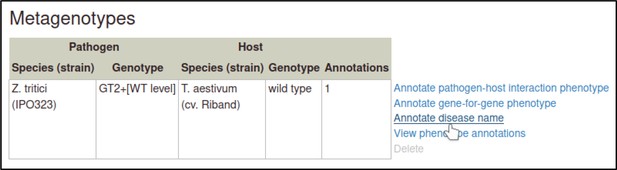
The Pathogen–Host Interaction Community Annotation Tool (PHI-Canto) provides the ‘Disease name’ annotation type, which is used to annotate a disease to a pathogen–host interaction.
These annotations highlight the fact that two different pathogens infecting different tissue types of the same host have been used in experiments within this publication. Disease name annotations are made on the Metagenotype Management page, via the ‘Annotate disease name’ link.

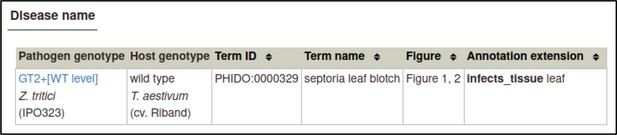
The curator can select a disease from a list of disease names provided by the Pathogen–Host Interactions database (PHI-base) Disease List (PHIDO).
For Z. tritici, the disease is septoria leaf blotch (PHIDO:0000329).
Disease name annotations also allow the host tissue infected to be specified. In this case, the tissue is the leaf (BTO:0000713).

The curator has the option to provide the figure number and additional comments.
In this case, the figure numbers are 1 and 2.
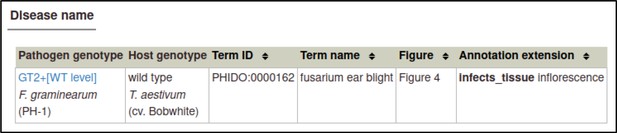
The same process can be followed to create the Disease name annotation for F. graminearum: the genotype is the wild-type GT2, the host cultivar is cv. Bobwhite, the disease is fusarium ear blight (PHIDO:0000162), the host tissue infected is the inflorescence (BTO:0000628), and the figure number is 4.
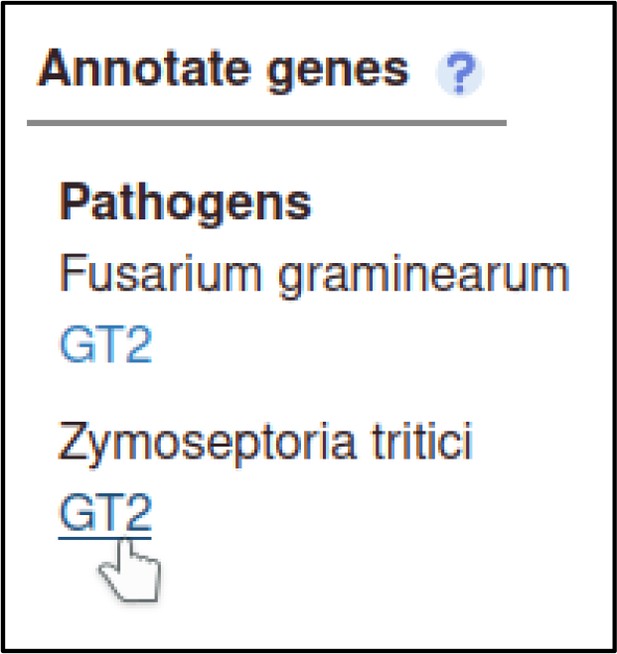
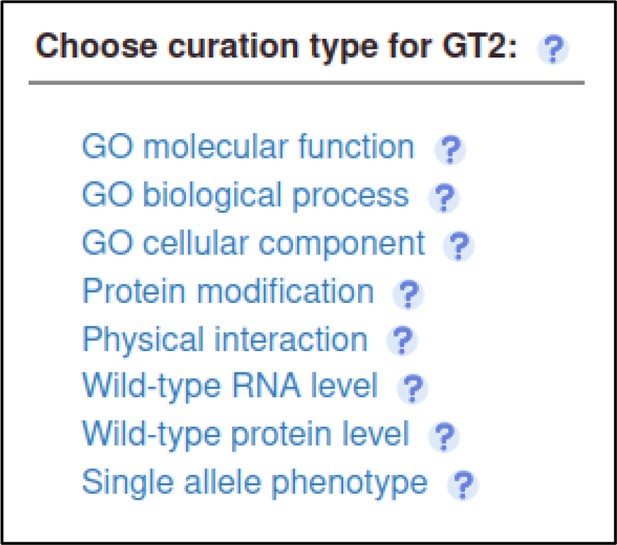
The Pathogen–Host Interaction Community Annotation Tool (PHI-Canto) also provides the ability to annotate biological processes, molecular functions, and cellular components associated with wild-type versions of genes, using terms from the Gene Ontology (GO).
In this publication, GT2 is described as having glycosyltransferase activity as its molecular function, so the curator can annotate this. Gene Ontology annotations are made by selecting the gene from the Curation Summary page.
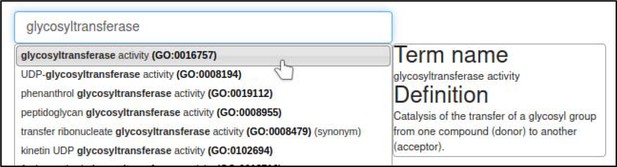
The curator selects the Gene Ontology (GO) Molecular Function annotation type and is prompted for a term from the Gene Ontology.
In this case, the correct term is glycosyltransferase activity (GO:0016757).

The curator must provide an evidence code from a controlled list specified by the Gene Ontology.
The appropriate evidence code in this case is a Traceable Author Statement in the publication.
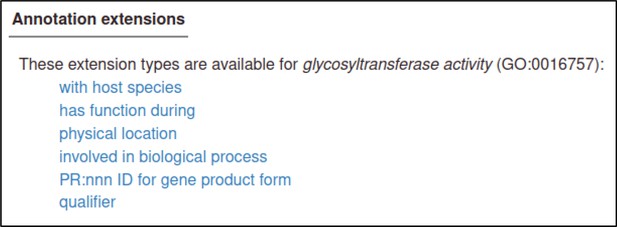
here are many annotation extensions available for Gene Ontology (GO) annotations, but in this case, none of them are applicable (or required), so the curator skips this step.

Figure numbers can be specified for Gene Ontology (GO) annotations: in this case, the relevant figure is Figure 3.

Once the curator has made all their annotations, the curation session is submitted to the PHI-base team for review.
The curator can use a text box to provide any information that is outside the scope of the curation process before finishing the submission process. Once the submission process is finished, the curation session can no longer be edited except by members of the Pathogen–Host Interactions database (PHI-base) team, who have the option to reactivate the session in case changes are required by the original curator.
Tables
Annotation types and annotation extensions in the Pathogen–Host Interaction Community Annotation Tool (PHI-Canto), grouped by the biological feature being annotated.
| Annotation type | Annotation extensions * | Annotation value |
|---|---|---|
| Annotation types for the gene biological feature † | ||
| Gene Ontology annotation | Gene Ontology term | |
| with host species | NCBI Taxonomy ID | |
| with symbiont species | NCBI Taxonomy ID | |
| Wild-type expression | PomBase Gene Expression ontology term | |
| during | Gene Ontology biological process term ‡ | |
| in presence of | Chemical entity (ChEBI ontology) | |
| tissue type | BRENDA Tissue Ontology term | |
| Annotation types for the genotype biological feature | ||
| Single species phenotype (Pathogen phenotype or Host phenotype) | PHIPO term (single-species phenotype branch) | |
| affected proteins | UniProtKB accession number (one for each affected protein) | |
| assayed RNA § | UniProtKB accession number | |
| assayed protein | UniProtKB accession number | |
| observed in organ | BRENDA Tissue Ontology term ¶ | |
| penetrance | Qualitative value (low, normal, high, complete) or quantitative value (percentage) | |
| severity | Qualitative value (low, normal, high, variable) or quantitative value (percentage) | |
| Annotation types for the metagenotype biological feature | ||
| Pathogen–host interaction phenotype or Gene-for-gene phenotype | PHIPO term (pathogen–host interaction phenotype branch) | |
| affected proteins | UniProtKB accession number (one for each affected protein) | |
| assayed protein | UniProtKB accession number | |
| assayed RNA | UniProtKB accession number | |
| compared to control metagenotype | Metagenotype ** | |
| extent of infectivity †† | PHIPO term | |
| gene-for-gene interaction ‡ ‡ | PHIPO Extension (PHIPO_EXT) ontology term | |
| host tissue infected | BRENDA Tissue Ontology term | |
| inverse gene-for-gene interaction ‡ ‡ | PHIPO Extension (PHIPO_EXT) ontology term | |
| outcome of interaction †† | PHIPO term | |
| penetrance | Qualitative value (low, normal, high, complete) or quantitative value (percentage) | |
| severity | Qualitative value (low, normal, high, variable) or quantitative value (percentage) | |
| Disease name | PHIDO term § § | |
| host tissue infected | BRENDA Tissue Ontology term | |
-
*
PHI-Canto uses 44 annotation extension (AE) relations, of which nine are unique to PHI-base, while the remaining 35 are shared with PomBase.
-
†
Additional AEs shared with PomBase for the gene annotation types are available in Supplementary file 2.
-
‡
Restricted to GO:0022403, GO:0033554, GO:0072690, GO:0051707 and their descendant terms.
-
§
AE relates to mRNA.
-
¶
Restricted to BTO:0001489, BTO:0001494, BTO:0001461 and their descendant terms.
-
**
Metagenotypes are selected from those already added to the curation session.
-
††
AE only applies to pathogen–host interaction phenotypes.
-
‡ ‡
AE only applies to gene-for-gene phenotypes.
-
§ §
Curated list of disease names.
Publications selected for trial curation using the Pathogen–Host Interaction Community Annotation Tool (PHI-Canto).
| Subject of publication | PMID | Publication title | Genotype * annotated with | Metagenotype † annotated with |
|---|---|---|---|---|
| Bacteria–human interaction | 28715477 ‡ | The RhlR quorum-sensing receptor controls Pseudomonas aeruginosa pathogenesis and biofilm development independently of its canonical homoserine lactone autoinducer. | Pathogen phenotype | unaffected pathogenicity, altered pathogenicity or virulence |
| Fungal–human interaction/novel antifungal target | 28720735 § | A nonredundant phosphopantetheinyl transferase, PptA, is a novel antifungal target that directs secondary metabolite, siderophore, and lysine biosynthesis in Aspergillus fumigatus and is critical for pathogenicity. | Pathogen phenotype | unaffected pathogenicity, altered pathogenicity or virulence |
| Secondary metabolite clusters required for pathogen virulence | 30459352 § | Phosphopantetheinyl transferase (Ppt)-mediated biosynthesis of lysine, but not siderophores or DHN melanin, is required for virulence of Zymoseptoria tritici on wheat. | Pathogen phenotype | unaffected pathogenicity, altered pathogenicity or virulence |
| Early acting virulence proteins | 29020037 §, ¶ | A conserved fungal glycosyltransferase facilitates pathogenesis of plants by enabling hyphal growth on solid surfaces. | Pathogen phenotype | altered pathogenicity or virulence |
| Mutualism interaction | 16517760 ** | Reactive oxygen species play a role in regulating a fungus-perennial ryegrass mutualistic interaction | Pathogen phenotype | mutualism |
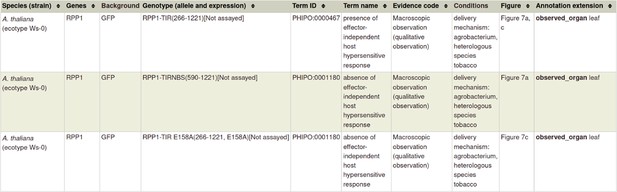
| First host targets of pathogen effectors | 31804478 §, †† | An effector protein of the wheat stripe rust fungus targets chloroplasts and suppresses chloroplast function. | N/A | altered pathogenicity or virulence a pathogen effector |
| Receptor decoys | 30220500 †† | Suppression of plant immunity by fungal chitinase-like effectors. | Pathogen phenotype | a pathogen effector |
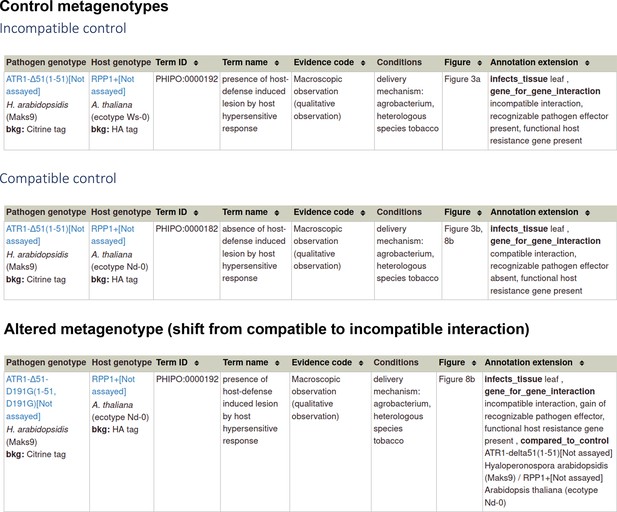
| R-Avr interactions | 20601497 ‡ ‡, § § | Activation of an Arabidopsis resistance protein is specified by the in planta association of its leucine-rich repeat domain with the cognate oomycete effector. | Host phenotype | a pathogen effector a gene-for-gene interaction |
| Fungal toxins required for virulence on plants | 22241993 ¶ ¶ | The cysteine rich necrotrophic effector SnTox1 produced by Stagonospora nodorum triggers susceptibility of wheat lines harboring Snn1. | N/A | a pathogen effector a gene-for-gene interaction (inverse) |
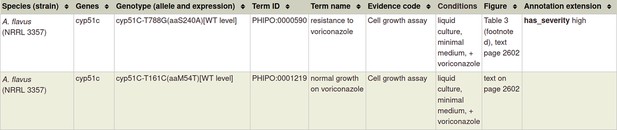
| Resistance to antifungal chemistries | 22314539 *** | The T788G mutation in the cyp51C gene confers voriconazole resistance in Aspergillus flavus causing aspergillosis. | Pathogen phenotype Pathogen chemistry phenotype | N/A |
-
*
Single species genotypes could be annotated with either a pathogen phenotype, a pathogen chemistry phenotype, or a host phenotype. Genotypes are annotated with in vitro or in vivo phenotypes from PHIPO, using either the Pathogen phenotype or Host phenotype annotation type workflow.
-
†
Metagenotype comprises of a pathogen and a host genotype in combination. Phenotypes from PHIPO can be annotated to metagenotypes using either the ‘Pathogen–Host Interaction Phenotype’ or ‘Gene-for-Gene Phenotype’ annotation type workflow.
-
‡
Example of curating 'unaffected pathogenicity' available in Appendix 1.
-
§
Example of curating 'altered pathogenicity or virulence' available in Appendix 1 and Appendix 2.
-
¶
Example of 'in vitro pathogen phenotype' available in Appendix 1.
-
**
Example of curating 'mutualism' available in Appendix 1. Although ‘mutualism interactions’ are generally out of scope for PHI-base, PHI-Canto can be used to curate these publications if required. In this study, the fungal gene mutation altered the interaction from mutualistic to antagonistic.
-
††
Example of curating 'a pathogen effector’ available in Appendix 1.
-
‡ ‡
Example of curating 'a gene-for-gene interaction' available in Appendix 1.
-
§ §
Example of 'in vivo host phenotype' available in Appendix 1.
-
¶ ¶
Example of curating 'an inverse gene-for-gene interaction' available in Appendix 1.
-
***
Example of 'in vitro pathogen chemistry phenotype' available in Appendix 1.
Issues encountered whilst curating ten example publications with the Pathogen–Host Interaction Community Annotation Tool (PHI-Canto).
| Curated feature | Problem description | Solution | Context in PHI-Canto | Example |
|---|---|---|---|---|
| Species strain | UniProtKB sequence information is commonly from a reference genome strain. This sequence may differ from the experimental strain curated in PHI-Canto. | Develop a selectable list of strains for curators to assign to the genotype (and metagenotype). | Strain selected after UniProtKB entry on gene entry page. Strain used within genotype creation. | URL1 All phenotype annotation examples in Appendix 1 contain a ‘strain name’ within the genotype/metagenotype. |
| Delivery mechanism | Pathogen–host interaction experiments use a wide array of mechanisms to deliver the treatment of choice (to cells, tissues, and host and non-host species) which are required for experimental interpretation. | Develop terms prefixed with ‘delivery mechanism’ in the Pathogen–Host Interaction Experimental Conditions Ontology (PHI-ECO). | Selection of experimental conditions whilst making a phenotype annotation to a metagenotype. | URL2 Examples in Appendix 1 PMID:20601497, PMID:31804478 and PMID:22241993. |
| Physical interaction | Physical interactions (i.e. protein–protein interactions) could only be annotated between proteins of the same species, so it was not possible to annotate interactions between a pathogen effector and its first host target. | Adapt the ‘Physical Interaction’ annotation type to store gene and species information from two organisms (instead of one). | Physical Interaction annotation type. | URL3 |
| Pathogen effector | There was no available ontology term to describe a ‘class’ pathogen effector (a ‘transferred entity from pathogen to host’), because effectors have heterogeneous functions (specific enzyme inhibitors, modulating host immune responses, and targeting host gene-silencing mechanisms). Effector is not a phenotype, and so did not fit into the Pathogen–Host Interaction Phenotype Ontology (PHIPO). | Develop new Gene Ontology (GO) biological process terms (and children), to group ‘effector-mediated’ processes. | GO Biological Process annotation on a pathogen gene. | URL4 Example in Appendix 1 PMID:31804478. |
| Wild-type control phenotypes | Natural sequence variation between strains of both pathogen and host organisms can alter the phenotypic outcome within an interaction. The wild-type metagenotype phenotype needs to be curated so that the phenotype of an altered metagenotype is informative. | Allow creation of metagenotypes containing wild-type genes. Develop a new annotation extension (AE) property ‘compared to control,’ used in annotation of altered metagenotypes. | Annotation of phenotypes and AEs to metagenotypes (using the ‘PHI phenotype’ or ‘Gene for gene phenotype’ annotation type). | URL5 Examples in Appendix 1 PMID:28715477, PMID:16517760, PMID:29020037, PMID:20601497, PMID:22241993. |
| Chemistry | How to record chemicals for resistance or sensitivity phenotypes. | Follow PomBase model to pre-compose PHIPO terms to include chemical names from the ChEBI ontology. | Annotation of phenotypes to single species genotypes. | URL4 Example in Appendix 1 PMID:22314539. |
| Gene for gene interactions | Complex gene-for-gene interactions within plant pathogen–host interactions required additional detail to describe the function of the pathogen and host genes within the metagenotype (including the specified strains). | Develop the additional metagenotype curation type ‘Gene for gene phenotype.’ Develop two new AEs, ‘gene_for_gene_interaction’ and ‘inverse gene_for_gene_interaction,’ using PHIPO_EXT terms describing three components of the interaction.* | Annotation of phenotypes and AEs to metagenotypes using the ‘Gene for gene phenotype’ annotation type. | URL4 Examples in Appendix 1 PMID:20601497 and PMID:22241993. |
| Nine high-level legacy terms (from PHI-base 4) | PHI-base should incorporate legacy data from PHI-base 4 into new PHI-base 5 gene-centric pages. | Maintain the nine high level terms as ‘tags’ within the new PHI-base 5 user interface. Develop mapping methods to enable this. | Three locations described in Supplementary file 3. | Urban et al., 2015 NAR (PMID:25414340). |
-
*
Namely, (i) the compatibility of the interaction (ii) the functional status of the pathogen gene, and (iii) the functional status of the host gene.
Automatically and manually curated types of data displayed in the gene-centric version 5 of the Pathogen–Host Interactions database (PHI-base).
| Data type | Data source |
|---|---|
| Metadata | |
| Entry Summary * | UniProtKB † |
| Pathogen species | NCBI Taxonomy † |
| Pathogen strain | PHI-base strain list |
| Host species | NCBI Taxonomy † |
| Host strain | PHI-base strain list |
| Publication | PubMed † |
| Phenotype annotation sections | |
| Pathogen–Host Interaction Phenotype | PHIPO ‡ pathogen–host interaction phenotype branch |
| Gene-for-Gene Phenotype | PHIPO pathogen–host interaction phenotype branch |
| Pathogen Phenotype | PHIPO single species phenotype branch |
| Host Phenotype | PHIPO single species phenotype branch |
| Other annotation sections | |
| Disease name | PHIDO |
| GO Molecular Function | GO § |
| GO Biological Process | GO |
| GO Cellular Component | GO |
| Wild-type RNA level ¶ | FYPO_EXT ** |
| Wild-type Protein level | FYPO_EXT |
| Physical Interaction | BioGRID †† |
| Protein Modification | PSI-MOD ‡ ‡ |
-
*
The Entry Summary section includes information on which gene is being displayed in the gene-centric results page. The UniProtKB accession number is used to automatically retrieve the name and function of the protein, plus any cross-referenced identifiers from Ensembl Genomes and NCBI GenBank. The section also displays the PHI-base 5 gene identifier (PHIG) and any of the high-level terms (Supplementary file 3) annotated to the gene.
-
†
Data from UniProtKB, NCBI Taxonomy, and PubMed are automatically retrieved, while all other data are manually curated.
-
‡
PHIPO is the Pathogen–Host Interaction Phenotype Ontology.
-
§
GO is the Gene Ontology.
-
¶
This relates to mRNA.
-
**
FYPO_EXT is the Fission Yeast Phenotype Ontology Extension.
-
††
BioGRID is the Biological General Repository for Interaction Datasets.
-
‡ ‡
PSI-MOD is the Human Proteome Organization (HUPO) Proteomics Standards Initiative (PSI) Protein Modifications Ontology.
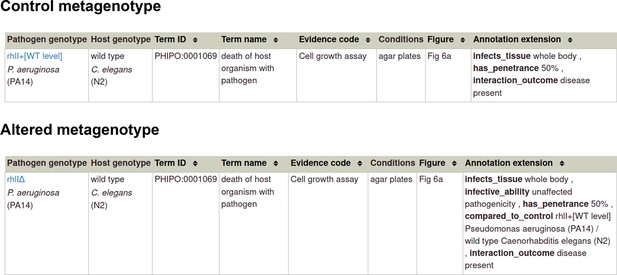
Annotation extensions (AE) summary for ‘unaffected pathogenicity’.
| AE name | Cardinality | Available terms |
|---|---|---|
| compared to control genotype | 0, 1 | Metagenotype identifier |
| extent of infectivity | 0, 1 | ‘unaffected pathogenicity’ |
| host tissue affected | 0, n | BRENDA Tissue Ontology term |
| outcome of interaction | 0, 1 | ‘disease present,’ ‘disease absent’ |
Annotation extensions (AE) summary for ‘altered pathogenicity or virulence’.
| AE name | Cardinality | Available terms |
|---|---|---|
| compared to control genotype | 0, 1 | Metagenotype identifier |
| extent of infectivity | 0, 1 | ‘loss of pathogenicity,’ ‘reduced virulence,’ ‘increased virulence’ |
| host tissue affected | 0, n | BRENDA Tissue Ontology term |
| outcome of interaction | 0, 1 | ‘disease present,’ ‘disease absent’ |
Annotation extensions (AE) summary for ‘mutualism’.
| AE name | Cardinality | Available terms |
|---|---|---|
| compared to control genotype | 0, 1 | Metagenotype identifier |
| extent of infectivity | 0, 1 | ‘mutualism present,’ ‘mutualism absent,’ ‘loss of mutualism’ |
| host tissue affected | 0, n | BRENDA Tissue Ontology term |
-
Note: The ‘Outcome of interaction’ AE is not relevant in this mutualism interaction.
Annotation extensions (AE) summary for ‘a pathogen effector’.
| AE name | Cardinality | Available terms |
|---|---|---|
| compared to control genotype | 0, 1 | Metagenotype identifier |
| extent of infectivity | 0, 1 | ‘unaffected pathogenicity,’ ‘loss of pathogenicity,’ ‘reduced virulence,’ ‘increased virulence’ |
| host tissue affected | 0, n | BRENDA Tissue Ontology term |
| outcome of interaction | 0, 1 | ‘disease present,’ ‘disease absent’ |
Annotation extensions (AE) summary for ‘a gene-for-gene interaction’.
| AE name | Cardinality | Available terms |
|---|---|---|
| compared to control genotype | 0, 1 | Metagenotype identifier |
| gene-for-gene phenotype | 0, 1 | ‘incompatible interaction, recognizable pathogen effector present, functional host resistance gene present’ ‘incompatible interaction, recognizable pathogen effector present, gain of functional host resistance gene’ ‘incompatible interaction, gain of recognizable pathogen effector, gain of functional host resistance gene’ ‘incompatible interaction, gain of recognizable pathogen effector, functional host resistance gene present’ ‘compatible interaction, recognizable pathogen effector present, functional host resistance gene absent’ ‘compatible interaction, recognizable pathogen effector absent, functional host resistance gene present’ ‘compatible interaction, recognizable pathogen effector present, compromised host resistance gene’ ‘compatible interaction, recognizable pathogen effector absent, functional host resistance gene absent’ ‘compatible interaction, recognizable pathogen effector absent, compromised functional host resistance gene’ ‘compatible interaction, compromised recognizable pathogen effector, functional host resistance gene present’ ‘metagenotype outcome overcome by external condition’ |
| host tissue affected | 0, n | BRENDA Tissue Ontology term |
Annotation extensions (AE) summary for ‘an inverse gene-for-gene interaction’.
| AE name | Cardinality | Available terms |
|---|---|---|
| compared to control genotype | 0, 1 | Metagenotype identifier |
| inverse gene-for-gene phenotype | 0, 1 | ‘compatible interaction, functional pathogen necrotrophic effector present, functional host susceptibility locus present’ ‘compatible interaction, functional pathogen necrotrophic effector present, gain of functional host susceptibility locus’ ‘compatible interaction, gain of functional pathogen necrotrophic effector, functional host susceptibility locus present’ ‘incompatible interaction, functional pathogen necrotrophic effector present, functional host susceptibility locus absent’ ‘incompatible interaction, functional pathogen necrotrophic effector absent, functional host susceptibility locus present’ ‘incompatible interaction, functional pathogen necrotrophic effector present, functional host susceptibility locus compromised’ ‘incompatible interaction, compromised functional pathogen necrotrophic effector, functional host susceptibility locus present’ ‘incompatible interaction, gain of functional pathogen necrotrophic effector, functional host susceptibility locus compromised’ ‘metagenotype outcome overcome by external condition’ |
| host tissue affected | 0, n | BRENDA Tissue Ontology term |
Annotation extensions (AE) summary for ‘curating single species phenotypes’.
| AE name | Cardinality | Available terms |
|---|---|---|
| affected proteins | 2 | UniProtKB accession number |
| assayed RNA | 0, 1 | UniProtKB accession number |
| assayed protein | 0, 1 | UniProtKB accession number |
| penetrance | 0, 1 | qualitative terms (‘high,’ ‘medium,’ ‘low,’ or ‘complete’) or a quantitative value (a percentage) |
| severity | 0, 1 | ‘high,’ ‘medium,’ ‘low,’ ‘variable severity’ |
| observed in organ | 0, 1 | BRENDA Tissue Ontology term |
Author checklist prior to publication.
| Point number | Point for the author to consider |
|---|---|
| 1 | Use the UniProtKB assigned gene name. Synonyms can be recorded in addition to the gene name. Prefix the gene name with the genus and species initials if the same genes from multiple species are used. |
| 2 | If reporting on a new (gene) sequence, submit your sequence to NCBI GenBank or the European Nucleotide Archive (ENA), then obtain an accession number prior to publication. Record this accession number within the manuscript. If reporting on a gene with an existing accession number, make sure this is reported in the manuscript. Please record the UniProtKB accession number for the protein of the gene, where available. Provide or use any existing informative allele or line designations for mutations and transgenes. |
| 3 | Provide a binomial species name for pathogen and host organisms, not just a common name. If possible, please also include NCBI Taxonomy IDs for the pathogen and host organisms at the rank of species. |
| 4 | Describe the tissue or organ in which the experimental observations were made (controlled language can be found in the BRENDA Tissue Ontology, see https://www.ebi.ac.uk/ols/ontologies/bto). |
| 5 | Describe any experimental techniques used, and accurately record any chemicals or reagents used. |
| 6 | When writing an article, try to keep the use of descriptive language as accurate and controlled as possible. For example, do not use ‘reduced pathogenicity’ or ‘loss of virulence,’ as these terms can be misleading: it would be more accurate to use 'reduced virulence’ and ‘loss of pathogenicity,’ respectively. Ideally, try to follow the terminology of an existing ontology: this will make the data easier to extract and reuse. Relevant ontologies include PHIPO and GO (https://www.ebi.ac.uk/ols/ontologies/phipo, https://www.ebi.ac.uk/ols/ontologies/go). |
| 7 | Document all the key information for the paper: do not rely on citing past papers for information on the pathogen used, or the strain used, and so on. |
| 8 | Think carefully when choosing keywords for your manuscript to ensure that the publication can be located by PHI-base’s keyword searches. One example of an ideal keyword is ‘pathogen-host interaction.’ |
| 9 | Record the provenance of the pathogen strain: for example, whether it is a lab strain or a field isolate, or if the strain was obtained from a stock center or as a gift from another lab. |
Additional files
-
Supplementary file 1
Mapping display name to relation name for Annotation Extensions in the Pathogen–Host Interaction Community Annotation Tool (PHI-Canto).
- https://cdn.elifesciences.org/articles/84658/elife-84658-supp1-v1.docx
-
Supplementary file 2
PomBase annotation extensions used in the Pathogen–Host Interaction Community Annotation Tool (PHI-Canto).
- https://cdn.elifesciences.org/articles/84658/elife-84658-supp2-v1.xlsx
-
Supplementary file 3
Pathogen–Host Interactions database (PHI-base) nine high-level term mapping to the Pathogen–Host Interaction Community Annotation Tool (PHI-Canto).
- https://cdn.elifesciences.org/articles/84658/elife-84658-supp3-v1.docx
-
Supplementary file 4
The Pathogen–Host Interaction Community Annotation Tool (PHI-Canto) species and strain lists for pathogens and hosts.
- https://cdn.elifesciences.org/articles/84658/elife-84658-supp4-v1.xlsx
-
Supplementary file 5
Mapping between strains in the Pathogen–Host Interactions database (PHI-base) and the Pathogen–Host Interaction Community Annotation Tool (PHI-Canto).
- https://cdn.elifesciences.org/articles/84658/elife-84658-supp5-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/84658/elife-84658-mdarchecklist1-v1.pdf
-
Source code 1
Main configuration file for the Pathogen–Host Interaction Community Annotation Tool (PHI-Canto).
This is the main configuration file for PHI-Canto. Much of the configuration is inherited from Canto, the original curation application from which PHI-Canto is derived. Lines containing custom configuration for PHI-Canto have been indicated with comments
- https://cdn.elifesciences.org/articles/84658/elife-84658-code1-v1.zip