Pynapple, a toolbox for data analysis in neuroscience
Figures
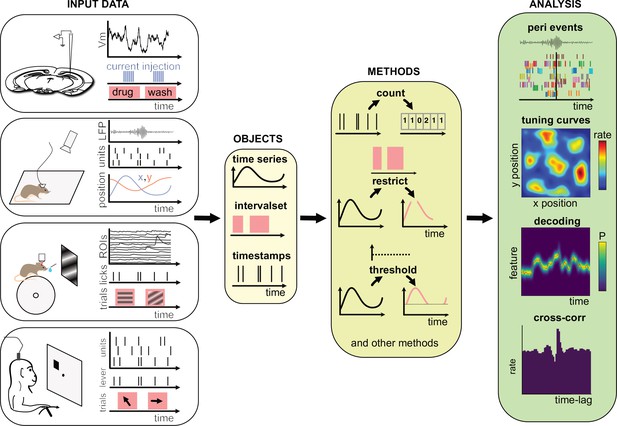
Data analysis with the Pynapple package.
Left, any type of input data can be loaded in a small number of core objects. For example (from top to bottom): intracellular recordings in slice during which current is injected and drug is applied to the bath solution; extracellular recordings in freely moving mice whose position is video-tracked; calcium imaging in head-fixed mice during presentation of different visual stimuli and delivery of precisely timed rewards; extracellular recordings in non-human primates during the execution of cognitive tasks. Middle, object-specific methods allow the user to perform a wide variety of basic operations and to manipulate the data manipulations. Right, at a higher level, the package contains a set of foundational analysis methods such as (from top to bottom) peri-event alignment of the data (top), 1- and 2D tuning curves, 1- and 2D decoding; auto- and cross-correlation of event times (e.g., action potentials). These methods depend only on a few, commonly used, external packages.
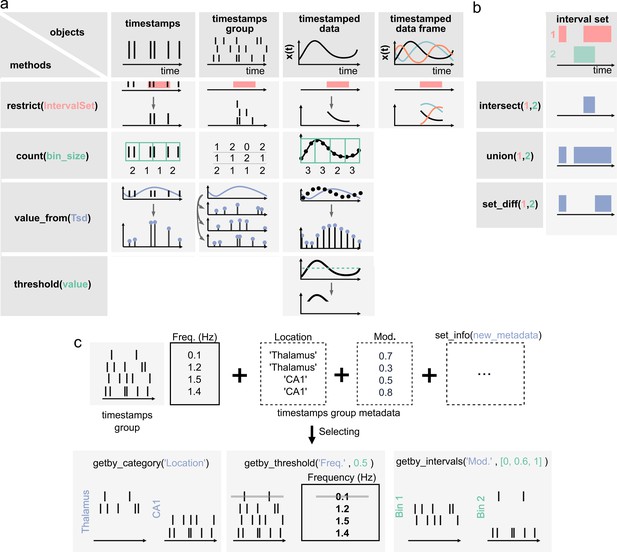
Core methods of the Pynapple objects.
(a) Methods of timestamps (Ts) and timestamped data (Tsd) objects. The same methods can be called for different objects, leading to qualitatively similar results. For example, object.restrict(intervalset) returns an object now defined on the intersection of its original time support and the input IntervalSet. Objects can be any of the timestamps and timestamped data objects. These methods can be called with only one argument, as shown here, since the default parameters are typically the same for most analyses. Yet the methods include additional arguments for more specific operations. (b) Logical operations on pairs of IntervalSet objects to compute (from top to bottom) the intersection, union, and difference between epochs. These operations are commonly used to analyze data during specific epochs in a combinatorial manner, such as ‘exploration period AND running speed is above 5 cm/s NOT left arm’. (c) Methods of TsGroup objects. Each timestamp is associated by default with its occurrence rate. Additional custom metadata such as recording location can be added. These metadata can then be used to select and filter timestamps using getby_category for discrete labels, getby_threshold, or getby_intervals for numerical values.
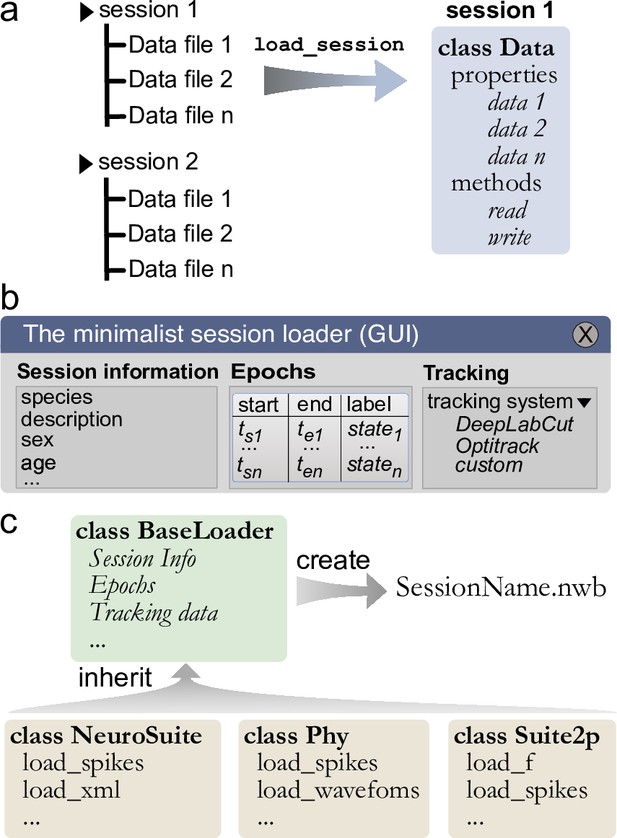
Built-in and customizable loading function for Pynapple.
(a) Data is originally organized as separate files in a folder. A built-in or custom-made load_session function is called to load the data into a Data class. (b) Data can be loaded through a customizable graphical user interface (GUI) to enter all relevant information regarding the experiment, for example animal strain, among others. The main epochs of the recording (e.g., behavioral states, stimuli category, etc.) can be loaded from standard tabular data files (such as CSV). Behavioral tracking data extracted from various common systems and saved as a CSV file can also be loaded. (c) Pynapple offers various built-in loaders for commonly used data formats, as well as a template to easily design a customizable loader to adapt to any other format or specific task design.
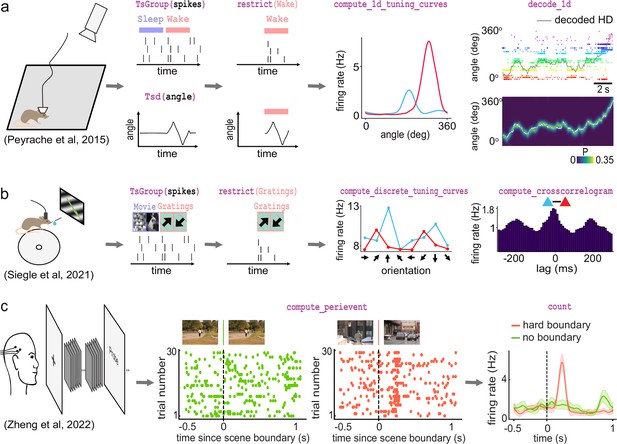
Examples of foundational analysis across various electrophysiological datasets using Pynapple.
(a) Analysis of an ensemble of head-direction cells. From left to right: data were collected in a freely moving mouse randomly foraging for food; all data are restricted to the wake epoch (i.e., during exploration); the tuning curve of two neurons relative to the animal’s head-direction; animal’s head-direction is decoded from the neuronal ensemble. Data from Peyrache et al., 2015a; Peyrache et al., 2015b. (b) Analysis of V1 neurons during visual stimulation. From left to right: the mouse was recorded while being head-fixed and presented with drifting gratings; spikes, stimulation, and epochs are shown; example tuning curves of two V1 neurons, showing their firing rates for different grating orientations; example cross-correlation between two V1 neurons, showing an oscillatory co-modulation at about 5 Hz during visual stimulation. Data from Siegle et al., 2021. (c) Analysis of medial temporal lobe neurons in human epileptic subjects. From left to right: subjects, implanted with hybrid deep electrodes, were shown a series of short clips; raster plot of a single neuron around continuous movie shot trials (green) and hard boundary trials, which are transitions between two unrelated movies (orange); peri-event neuronal firing rate for both trial types. Data from Zheng et al., 2022. Images in panels b and c are from Olmos and Kingdom, 2004. The analysis code used to generate this figure can be found on the Pynapple Organization GitHub repository: https://github.com/pynapple-org/pynapple-paper-2023, swh:1:rev:2603975ce421a02a30b82a05a2c1bda810246f9d; (Viejo, 2023a).
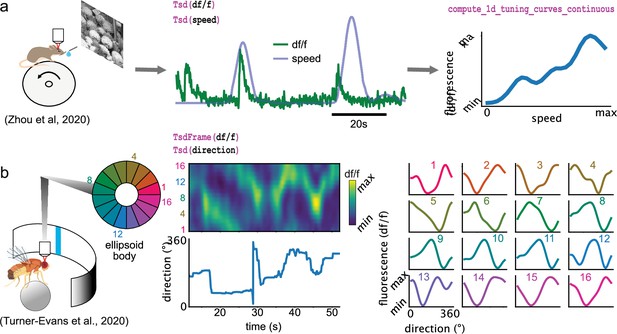
Examples of foundational analysis across various calcium imaging datasets using Pynapple.
(a) Analysis of a V1 neuron during visual stimulation. From left to right: the mouse was recorded while being head-fixed on a running wheel and presented with natural scene movies; fluorescence traces from a preprocessed region of interest and running speed are loaded; continuous tuning curve is directly obtained from fluorescence and speed. Data from Zhou et al., 2020. Image is from Olmos and Kingdom, 2004. (b) Analysis of neuronal activity in the fly central complex. From left to right: a Drosophila melanogaster is tethered to a calcium imaging setup while the position of a vertical bar is in closed loop with the fly’s movements on a ball; calcium activity in the ellipsoid body is divided into 16 wedges; example fluorescence trace and direction of the fly. Tuning curves are obtained as in (a), with the direction as feature. Data from Turner-Evans et al., 2020.

The Pynapple collaborative data analysis repository (Pynacollada) environment.
Unlike Pynapple, which is designed for long-term stability, Pynacollada is a repository of project-oriented libraries. This way, the community can collaborate on constantly evolving data analysis code without affecting the functionality of the core pipeline. Each project should include a script that can be called for specific functions and/or Jupyter notebooks to showcase the use of the code, as well as proper documentation. Pynacollada already includes several libraries and/or tutorials, including but not limited to: (1) a tutorial on manifold analysis, covering how to project neuronal data on low-dimensional subspace using various machine learning techniques; (2) a library for oscillation detection in local field potentials, which takes raw broadband traces as inputs and outputs IntervalSet objects corresponding to the start and end times of oscillation bouts.





