Coevolution of the CDCA7-HELLS ICF-related nucleosome remodeling complex and DNA methyltransferases
Figures
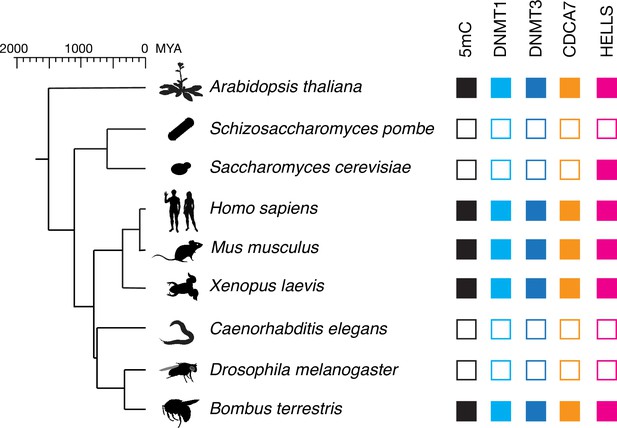
CDCA7 is absent from model organisms with undetectable genomic 5mC.
Filled squares and open squares indicate presence and absence of an orthologous protein(s), respectively. CDCA7 homologs are absent from model organisms where DNMT1, DNMT3 and 5mC on genomic are absent.
-
Figure 1—source data 1
Lists of proteins and species used in this study.
Tab1, Full list. The list contains species names, their taxonomies, Genbank accession numbers of proteins, PMID of references supporting the 5mC status, and genome sequence assembly statistics. ND; not detected. DNMT5 proteins shown in red lack the Snf2-like ATPase domain. UHRF1 proteins shown in red lack the Ring-finger E3 ubiquitin-ligase domain. CDCA7 proteins shown in red indicate ambiguous annotation as described in the main text. CDCA7 orthologs that contain additional conserved domains found by NCBI CD-search were shown in light blue. Tab2, Full list 2. The list is used to make presence (1) or absence (0) list. Tab3 Ecdysozoa CoPAP. List of presence/absence annotations for Ecdysozoa species used for CO-PAP analysis. Tab4 Full CoPAP1. List of presence/absence data annotations for the panel of all 180 species used for CO-PAP analysis. Fungal CDCA7F proteins with class II zf-4CXXC_R1 are included in CDCA7. Tab5 Full CoPAP2. List of presence/absence data annotations for the panel of all 180 species used for CO-PAP analysis. Fungal CDCA7F proteins are included in class II zf-4CXXC_R1. Tab6 Full clustering. Table used for clustering analysis. Tab7 Metazoan invertebrates. Table used for clustering analysis for metazoan invertebrates. Tab7 No 5mC list. List of species where absence of genomic 5mC has been experimentally shown.
- https://cdn.elifesciences.org/articles/86721/elife-86721-fig1-data1-v1.xlsx
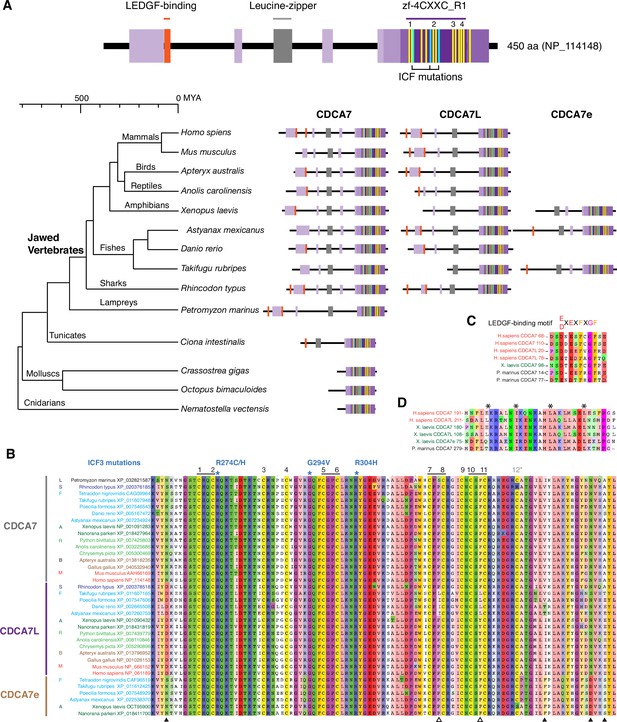
CDCA7 paralogs in vertebrates.
(A) Schematics of vertebrate CDCA7 primary sequence composition, based on NP_114148. Yellow lines and light blue lines indicate positions of evolutionary conserved cysteine residues and residues that are mutated in ICF patients, respectively. (B) Sequence alignment of the zf-4CXXC_R1 domain of vertebrate CDCA7-family proteins. White arrowheads; amino residues unique in fish CDCA7L. Black arrowheads; residues that distinguish CDCA7L and CDCA7e from CDCA7. (C) Sequence alignment of LEDGF-binding motifs. (D) Sequence alignment of the conserved leucine-zipper.
-
Figure 2—source data 1
Multiple sequence alignment of zf-4CXXC_R1 domains.
The zf-4CXXC_R1 domains were aligned by MUSCLE v5.
- https://cdn.elifesciences.org/articles/86721/elife-86721-fig2-data1-v1.zip
-
Figure 2—source data 2
An IQ-TREE result of the consensus phylogenetic tree generation of zf-4CXXC_R1 containing proteins.
Figure 2—source data 2 was used for the analysis by IQ-TREE.
- https://cdn.elifesciences.org/articles/86721/elife-86721-fig2-data2-v1.txt
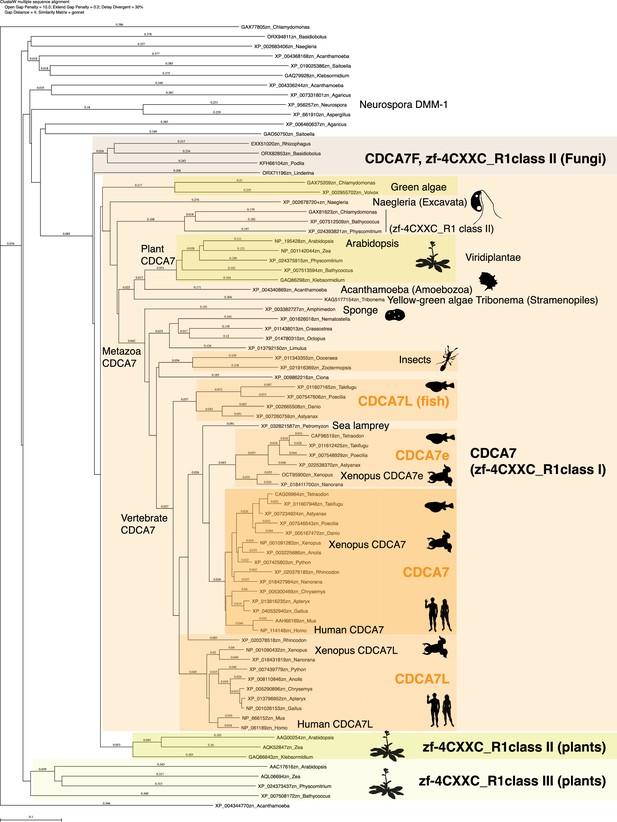
Evolutionary conservation of CDCA7-family proteins and other zf-4CXXC_R1-containig proteins.
Amino acid sequences of zf-4CXXC_R1 domain from indicated species were aligned with CLUSTALW. A phylogenetic tree of this alignment is shown. Genbank accession numbers of analyzed sequences are indicated. The tree topology was largely consistent with a tree generated by IQ-TREE based on an alignment using Muscle (Figure 2—source data 1 and Figure 2—source data 2).
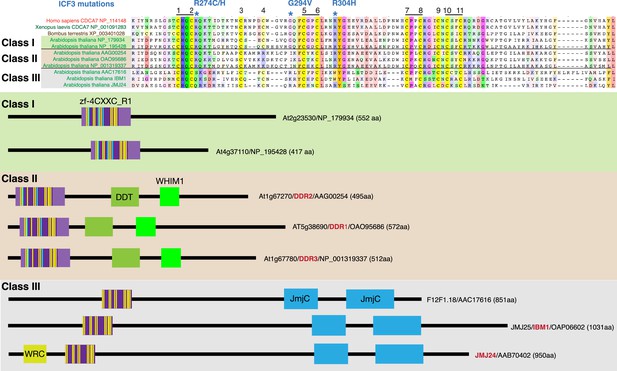
CDCA7 homologs and other zf-4CXXC_R1-containing proteins in Arabidopsis.
Top; alignments of the zf-4CXXC_R1 domain found in Arabidopsis thaliana. Bottom; domain structure of the three classes of zf-4CXXC_R1-containing proteins in Arabidopsis.
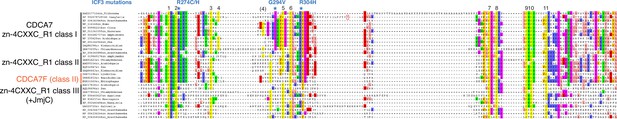
Sequence alignment and classification of zf-4CXXC_R1 domains across eukaryotes.
CDCA7 orthologs are characterized by the class I zf-4CXXC_R1 domain, where eleven cysteine residues and three residues mutated in ICF patients are conserved. Class II zf-4CXXC_R1 domain is similar to class I except that ICF-associated glycine (G294 in human) is substituted. Class III is zf-4CXXC_R domain with more substitutions at the ICF-associated residues (R274 and/or G294). Proteins that also contain JmjC domain (sequence not shown here) are indicated. Note that codon frame after the stop codon (an asterisk in a magenta box) of Naegleria XP_002678720 encodes a peptide sequence that aligns well with human CDCA7, indicating that the apparent premature termination of XP_002678720 is likely caused by a sequencing or annotation error.
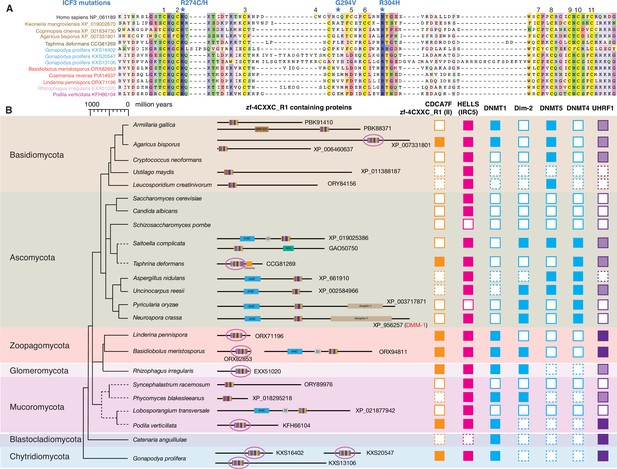
Evolutionary conservation of CDCA7F, HELLS and DNMTs in fungi.
(A) Sequence alignment of fungi-specific CDCA7F with class II zf-4CXXC_R1 sequences. (B) Domain architectures of zf-4CXXC_R1-containg proteins in fungi. The class II zf-4CXXC_R1 domain is indicated with purple circles. Squares with dotted lines indicate preliminary genome assemblies. Opaque boxes of UHRF1 indicate homologs that harbor the SRA domain but not the RING-finger domain.
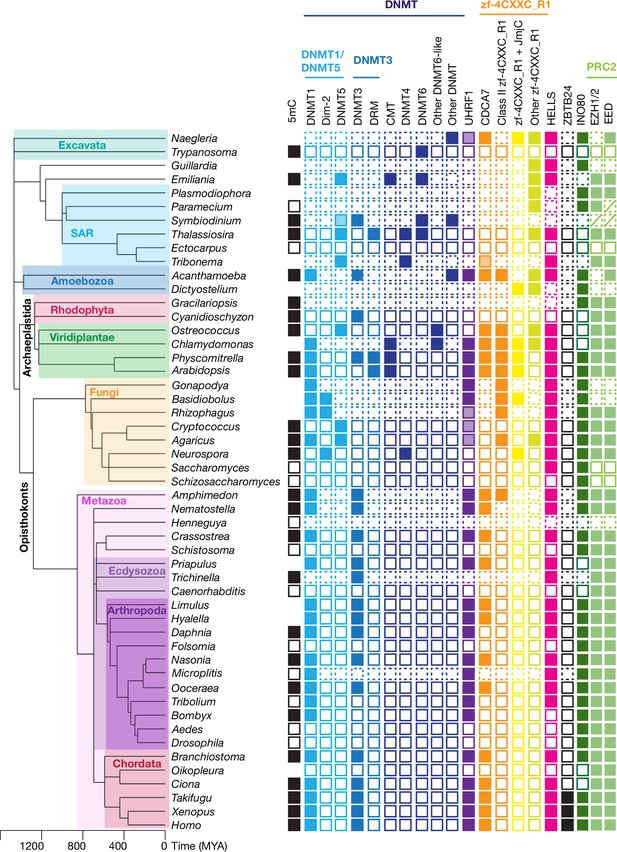
Evolutionary conservation of CDCA7, HELLS, and DNMTs.
The phylogenetic tree was generated based on Timetree 5 (Kumar et al., 2022). Filled squares and open squares indicate presence and absence of an orthologous protein(s), respectively. Squares with dotted lines imply preliminary-level genome assemblies. Squares with a diagonal line; Paramecium EED was functionally identified (Miró-Pina et al., 2022), but not by the sequence-based search in this study; homologs of EZH1/2 and EED were identified in Symbiodinium sp. KB8 but not in Symbiodinium microadriaticum (Figure 1—source data 1). An opaque box of DNMT5 in Symbiodinium indicates a homolog that does not contain the ATPase domain, which is commonly found in DNMT5 family proteins. Opaque boxes of UHRF1 indicate homologs that harbor the SRA domain but not the RING-finger domain. Full set of analysis on the panel of 180 eukaryote species is shown in Figure 5—figure supplement 1 and Figure 1—source data 1. Genbank accession numbers of each protein and PMID numbers of published papers that report presence or absence of 5mC are reported in Figure 1—source data 1.
-
Figure 5—source data 1
Multiple sequence alignment of the SNF2 ATPase domains of HELLS homologs and other SNF2-family proteins.
The SNF2 ATPase domains of HELLS and other SNF2-family proteins after removing the variable linker regions were aligned by MUSCLE v5.
- https://cdn.elifesciences.org/articles/86721/elife-86721-fig5-data1-v1.zip
-
Figure 5—source data 2
An IQ-TREE result of the consensus phylogenetic tree generation of HELLS homologs and other SNF2-family proteins.
Figure 5—source data 1 was used for the analysis by IQ-TREE.
- https://cdn.elifesciences.org/articles/86721/elife-86721-fig5-data2-v1.txt
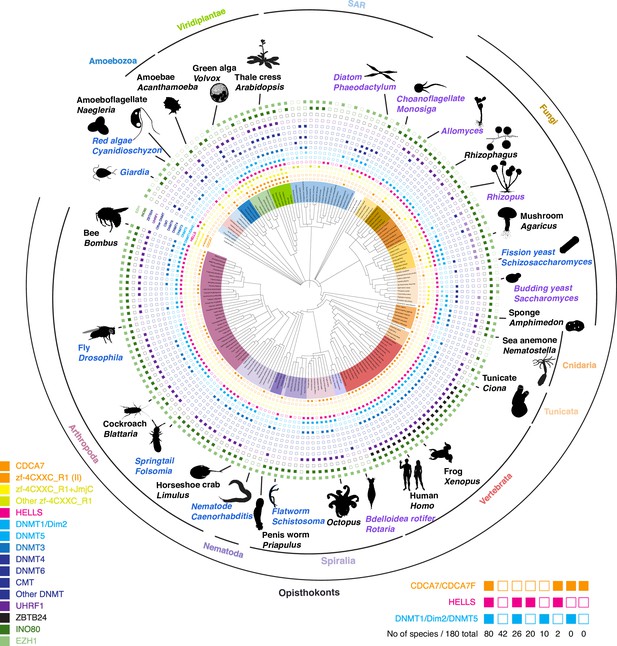
Evolutionary conservation of CDCA7, HELLS, and DNMTs.
Presence and absence of each annotated proteins in the panel of 180 eukaryote species is marked as filled and blank boxes. The phylogenetic tree was generated by iTOL, based on NCBI taxonomy by phyloT. Bottom right; summary of combinatory presence or absence of CDCA7 (including fungal CDCA7F containing class II zf-4CXXC_R1), HELLS, and maintenance DNA methyltransferases DNMT1/Dim-2/DNMT5. Supporting information including Genbank accession numbers are listed in Figure 1—source data 1.
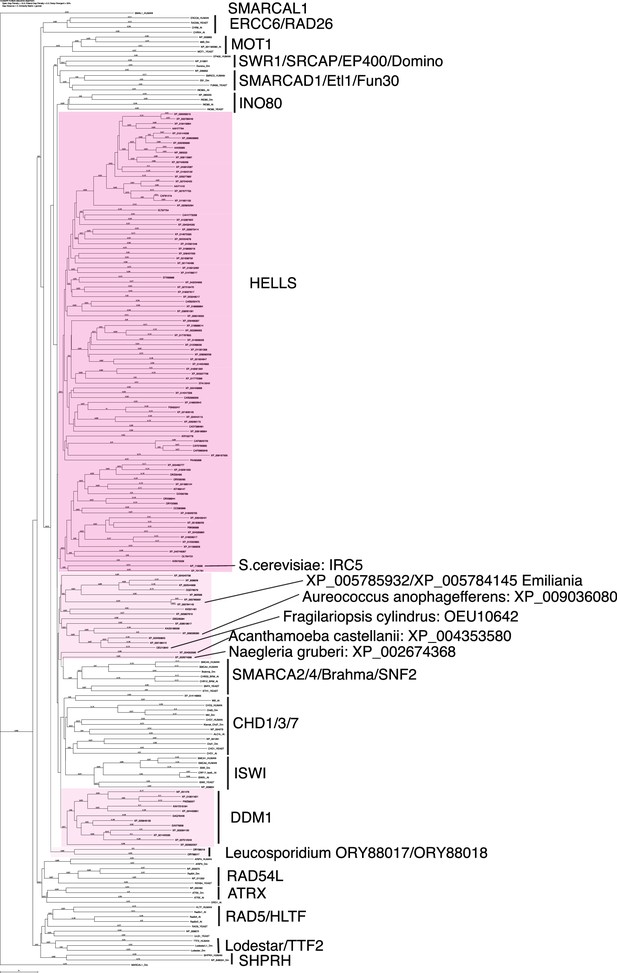
Phylogenetic tree of HELLS and other SNF2 family proteins.
Amino acid sequences of full-length HELLS proteins from the panel of 180 eukaryote species listed in Figure 1—source data 1 were aligned with full length sequences of other SNF2 family proteins with CLUSTALW. A phylogenetic tree of this alignment is shown. Genbank accession numbers of analyzed sequences are indicated.
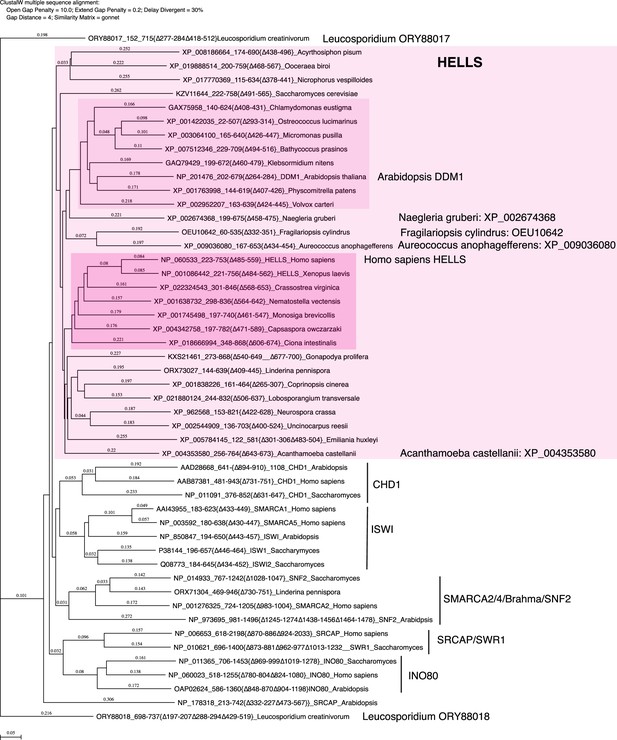
Phylogenetic tree of the SNF2-domain.
Amino acid sequences of SNF2-doman without variable insertions from representative HELLS and DDM1-like proteins from Figure 3 were aligned with the corresponding domain of other SNF2 family proteins with CLUSTALW. A phylogenetic tree of this alignment is shown. Genbank accession numbers of analyzed sequences are indicated. The tree topology was largely consistent with a tree generated by IQ-TREE based on an alignment using Muscle (Figure 5—source data 1 and Figure 5—source data 2).
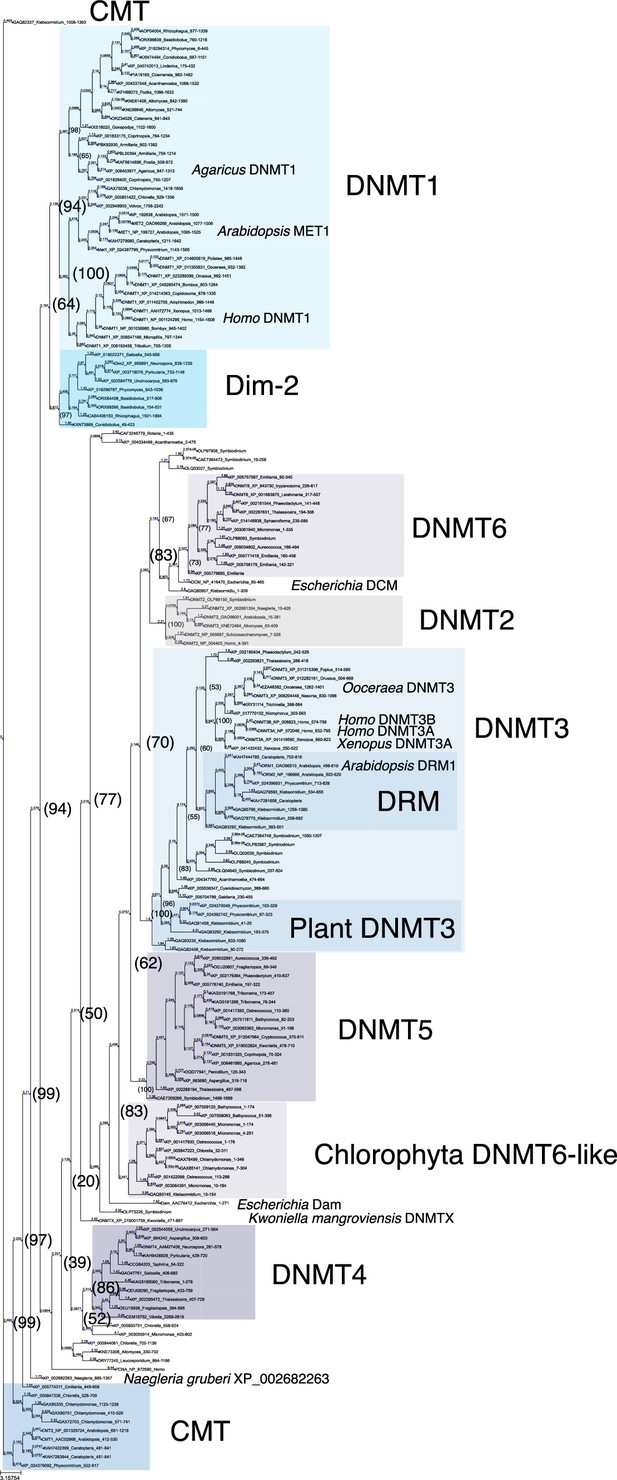
Phylogenetic tree of DNMT proteins.
DNA methyltransferase domain of DNMT proteins across eukaryotes (Figure 1—source data 1, excluding majority of those from Metazoa), the Escherichia coli DNA methylases DCM and Dam, and Homo sapiens PCNA as an outlier sequence, were aligned with Muscle, and a consensus phylogenetic tree was constructed from 1000 bootstrap trees using IQ-TREE. Branch lengths are optimized by maximum likelihood on original alignment. Numbers in parentheses are bootstrap supports (%).
-
Figure 5—figure supplement 4—source data 1
Multiple sequence alignment of DNA methyltransferase domains for Figure 5—figure supplement 4.
DNMT domains from various DNMTs were aligned by MUSCLE v5.
- https://cdn.elifesciences.org/articles/86721/elife-86721-fig5-figsupp4-data1-v1.zip
-
Figure 5—figure supplement 4—source data 2
An IQ-TREE result of the consensus phylogenetic tree generation of DNMTs for Figure 5—figure supplement 4.
Figure 5—figure supplement 4—source data 1 was used for the analysis by IQ-TREE.
- https://cdn.elifesciences.org/articles/86721/elife-86721-fig5-figsupp4-data2-v1.txt
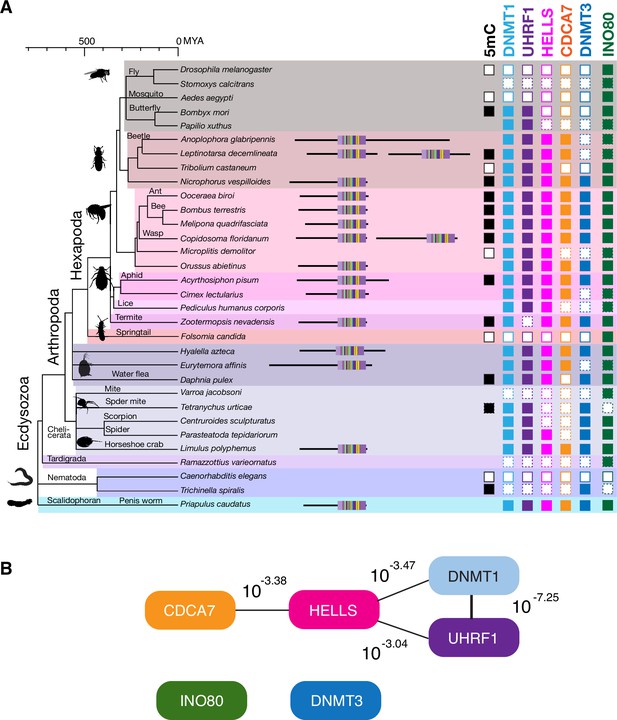
Coevolution of CDCA7, HELLS, UHRF1, and DNMT1 in Ecdysozoa.
(A) Presence (filled squares) /absence (open squares) patterns of indicated proteins and genomic 5mC in selected Ecdysozoa species. Squares with dotted lines imply preliminary-level genome assemblies. Domain architectures of CDCA7 proteins with a zf-4CXXC_R1 domain are also shown. (B) CoPAP analysis of 50 Ecdysozoa species. Presence/absence patterns of indicated proteins during evolution were analyzed. List of species are shown in Figure 1—source data 1. Phylogenetic tree was generated by amino acid sequences of all proteins shown in Figure 1—source data 1. The number indicates the p-values.
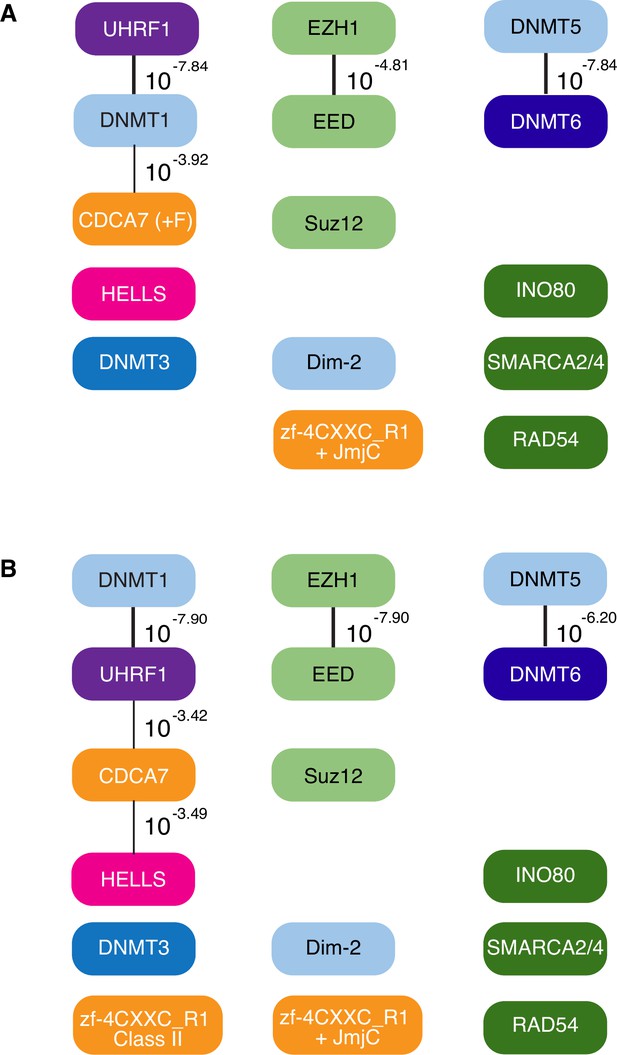
CoPAP analysis of CDCA7, HELLS, and DNMTs in eukaryotes.
CoPAP analysis of 180 eukaryote species. Presence and absence patterns of indicated proteins during evolution were analyzed. List of species are shown in Figure 1—source data 1 (A, Tab4. Full CoPAP1; B, Tab5. Full CoPAP2). Fungal CDCA7F proteins are included in CDCA7 and zf-4CXXC_R1 class II in A and B, respectively. Phylogenetic tree was generated by amino acid sequences of all proteins shown in Figure 1—source data 1. The number indicates the p-values.
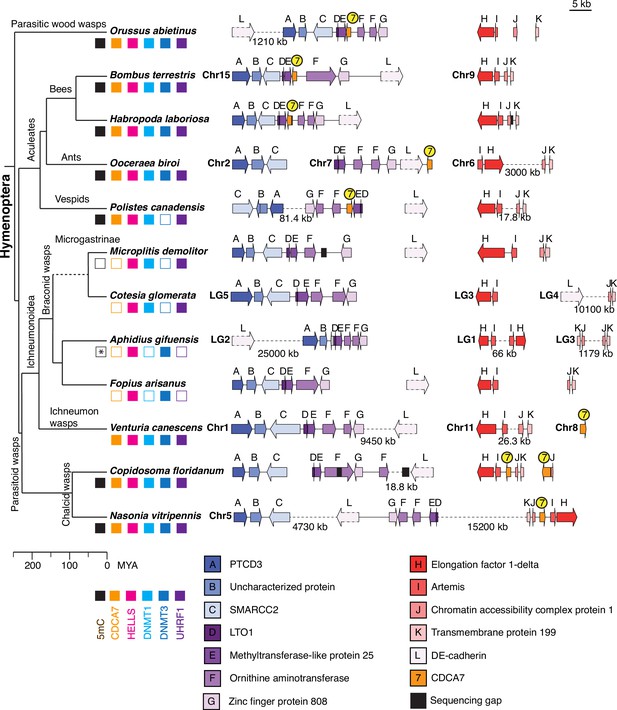
Synteny of Hymenoptera genomes adjacent to CDCA7 genes.
Genome compositions around CDCA7 genes in Hymenoptera insects are shown. For genome with annotated chromosomes, chromosome numbers (Chr) or linkage group numbers (LG) are indicated at each gene cluster. Gene clusters without chromosome annotation indicate that they are within a same scaffold or contig. Gene locations within each contig are listed in Figure 7—source data 1. Dash lines indicate the long linkages not proportionally scaled in the figure. Due to their extraordinarily long sizes, DE-cadherin genes (L) are not scaled proportionally. Presence and absence of 5mC, CDCA7, HELLS, DNMT1, DNMT3, and UHRF1 in each genome is indicated by filled and open boxes, respectively. Absence of 5mC in Aphidus gifuensis (marked with an asterisk) is deduced from the study in Aphidius ervi (Bewick et al., 2017b), which has an identical presence/absence pattern of the listed genes (Figure 7—source data 2).The phylogenetic tree is drawn based on published analysis (Li et al., 2021; Peters et al., 2017) and TimeTree.
-
Figure 7—source data 1
Summary of Hymenoptera genome location for Figure 7.
- https://cdn.elifesciences.org/articles/86721/elife-86721-fig7-data1-v1.xlsx
-
Figure 7—source data 2
Lists of proteins in Hymenoptera species supporting Figure 7.
- https://cdn.elifesciences.org/articles/86721/elife-86721-fig7-data2-v1.xlsx
Tables
| Reagent type(species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Software, algorithm | MacVector | MacVector, Inc | Version 16–18 | |
| Software, algorithm | Muscle | https://www.drive5.com/muscle/ | Muscle5.1 | |
| Software, algorithm | IQ-TREE | http://www.iqtree.org/ | Version 2.0.3 and 2.2.2.6 | |
| Software, algorithm | Timetree | http://www.timetree.org/ | Version 5 | |
| Software, algorithm | phyloT | https://phylot.biobyte.de/ | Version 2 | |
| Software, algorithm | iTOL | https://itol.embl.de/ | Version 6 | |
| Software, algorithm | CoPAP | http://copap.tau.ac.il/source.php | ||
| Software, algorithm | ETE Toolkit | http://etetoolkit.org/ | ||
| Software, algorithm | Jalview | https://www.jalview.org/ | Version 2.22.2.7 |





