Identifying metabolic features of colorectal cancer liability using Mendelian randomization
Figures

Study design.
First, linear regression models were used to examine the relationship between genetic susceptibility to adult colorectal cancer (CRC) and circulating metabolites measured in the Avon Longitudinal Study of Parents and Children (ALSPAC) participants at age 8 y, 16 y, 18 y, and 25 y. Next, we performed a reverse Mendelian randomization analysis to identify metabolites influenced by CRC susceptibility in an independent population of adults. Finally, we performed a conventional (forward) Mendelian randomization analysis of circulating metabolites on CRC to identify metabolites causally associated with CRC risk. Consistent evidence across all three methodological approaches was interpreted to indicate a causal role for a given metabolite in CRC aetiology.

Associations of genetic liability to adult colorectal cancer (based on a 72 single-nucleotide polymorphism [SNP] genetic risk score) with clinically validated metabolic traits at different early life stages among the Avon Longitudinal Study of Parents and Children (ALSPAC) offspring (age 8 y [N = 4767], 16 y [N = 2930], 18 y [N = 2613], and 25 y [N = 2559]).
Estimates shown are beta coefficients representing the SD difference in metabolic trait per doubling of genetic liability to colorectal cancer (purple, 8 y; turquoise, 16 y; red, 18 y; black, 25 y). Filled point estimates are those that pass a Benjamini–Hochberg FDR multiple-testing correction (FDR < 0.05).
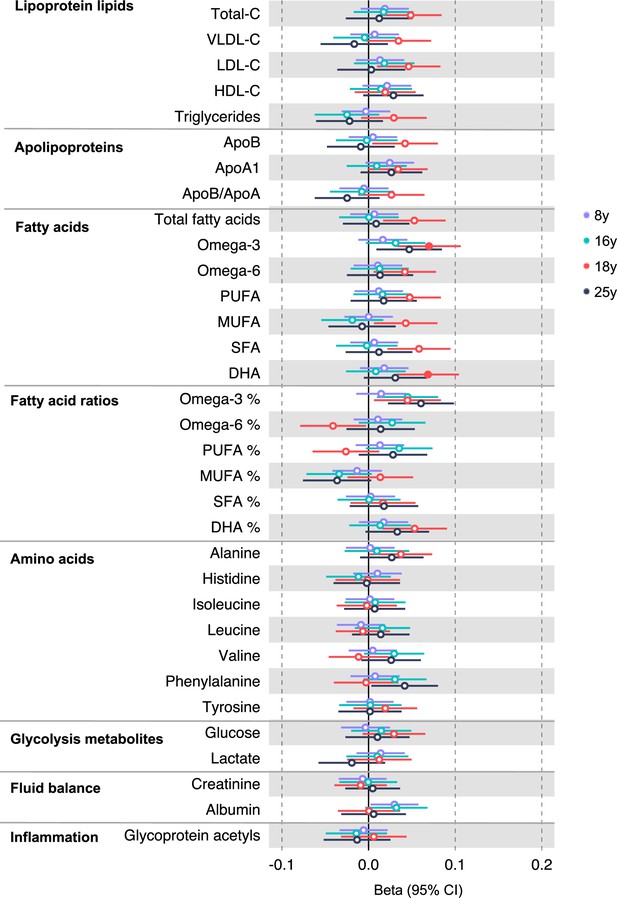
Associations of genetic liability to adult colon cancer with clinically validated metabolic traits at different early life stages among the Avon Longitudinal Study of Parents and Children (ALSPAC) offspring (age 8 y, 16 y, 18 y, and 25 y).
Estimates shown are beta coefficients representing the SD difference in metabolic trait per doubling of genetic liability to colon cancer (purple, 8 y; turquoise, 16 y; red, 18 y; black, 25 y). Filled point estimates are those that pass a Benjamini–Hochberg FDR multiple-testing correction (FDR < 0.05).
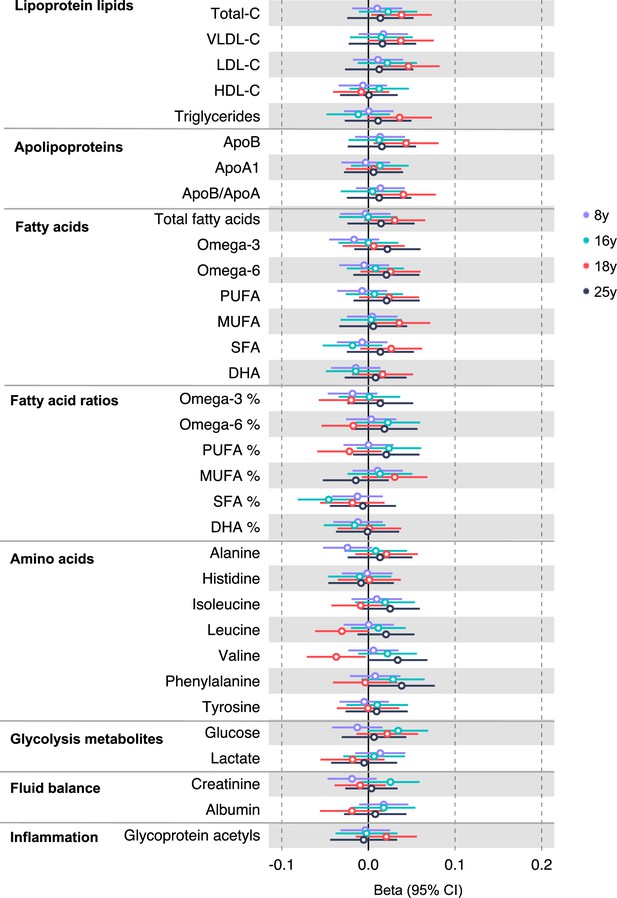
Associations of genetic liability to proximal colon cancer with clinically validated metabolic traits at different early life stages among the Avon Longitudinal Study of Parents and Children (ALSPAC) offspring (age 8 y, 16 y, 18 y, and 25 y).
Estimates shown are beta coefficients representing the SD difference in metabolic trait per doubling of genetic liability to proximal colon cancer (purple, 8 y; turquoise, 16 y; red, 18 y; black, 25 y). Filled point estimates are those that pass a Benjamini–Hochberg FDR multiple-testing correction (FDR < 0.05).
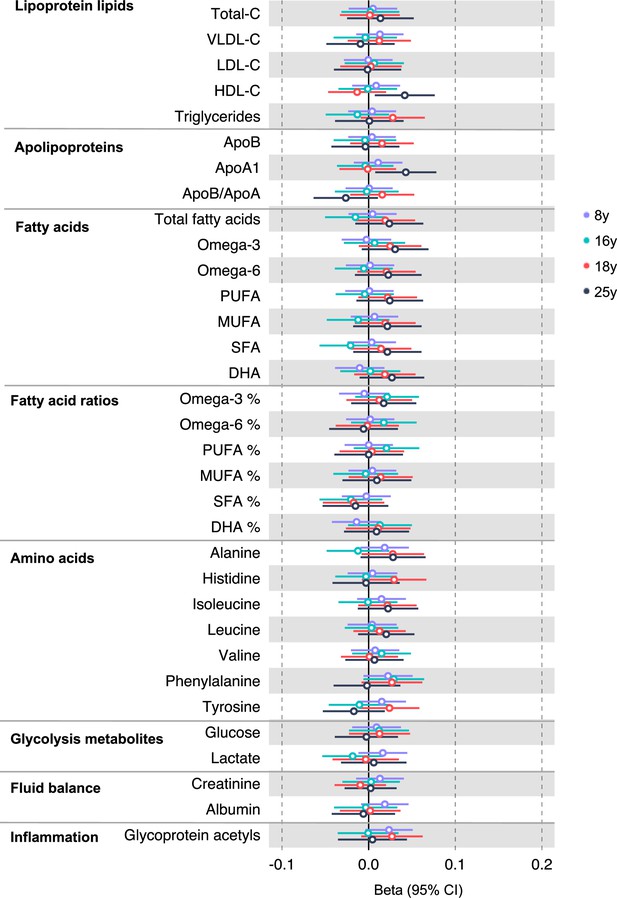
Associations of genetic liability to distal colon cancer with clinically validated metabolic traits at different early life stages among the Avon Longitudinal Study of Parents and Children (ALSPAC) offspring (age 8 y, 16 y, 18 y, and 25 y).
Estimates shown are beta coefficients representing the SD difference in metabolic trait per doubling of genetic liability to distal colon cancer (purple, 8 y; turquoise, 16 y; red, 18 y; black, 25 y). Filled point estimates are those that pass a Benjamini–Hochberg FDR multiple-testing correction (FDR < 0.05).
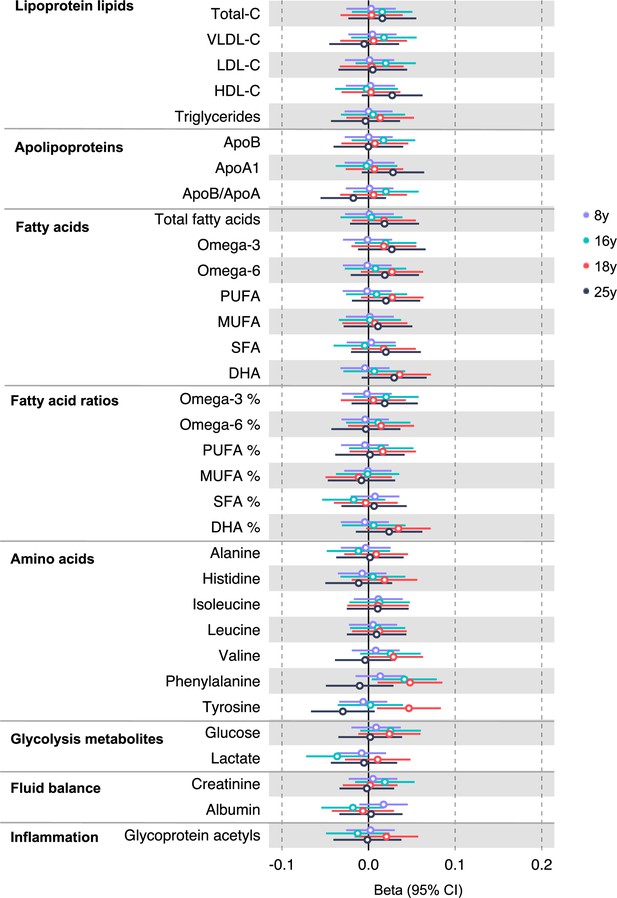
Associations of genetic liability to rectal cancer with clinically validated metabolic traits at different early life stages among the Avon Longitudinal Study of Parents and Children (ALSPAC) offspring (age 8 y, 16 y, 18 y, and 25 y).
Estimates shown are beta coefficients representing the SD difference in metabolic trait per doubling of genetic liability to rectal cancer (purple, 8 y; turquoise, 16 y; red, 18 y; black, 25 y). Filled point estimates are those that pass a Benjamini–Hochberg FDR multiple-testing correction (FDR < 0.05).
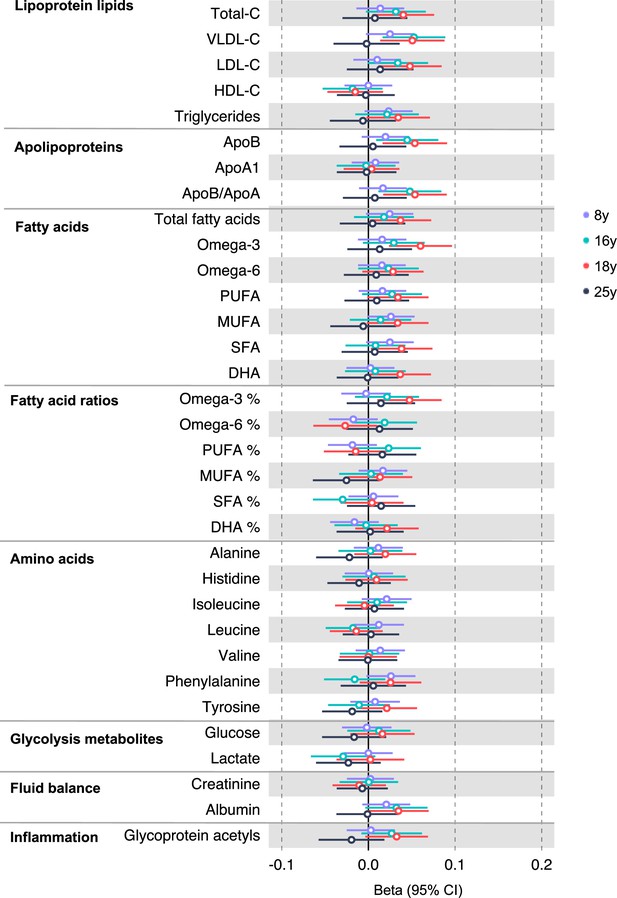
Associations of genetic liability to adult colorectal cancer (excluding rs174533) with clinically validated metabolic traits at different early life stages among the Avon Longitudinal Study of Parents and Children (ALSPAC) offspring (age 8 y, 16 y, 18 y, and 25 y).
Estimates shown are beta coefficients representing the SD difference in metabolic trait per doubling of genetic liability to colorectal cancer (purple, 8 y; turquoise, 16 y; red, 18 y; black, 25 y). Filled point estimates are those that pass a Benjamini–Hochberg FDR multiple-testing correction (FDR < 0.05).
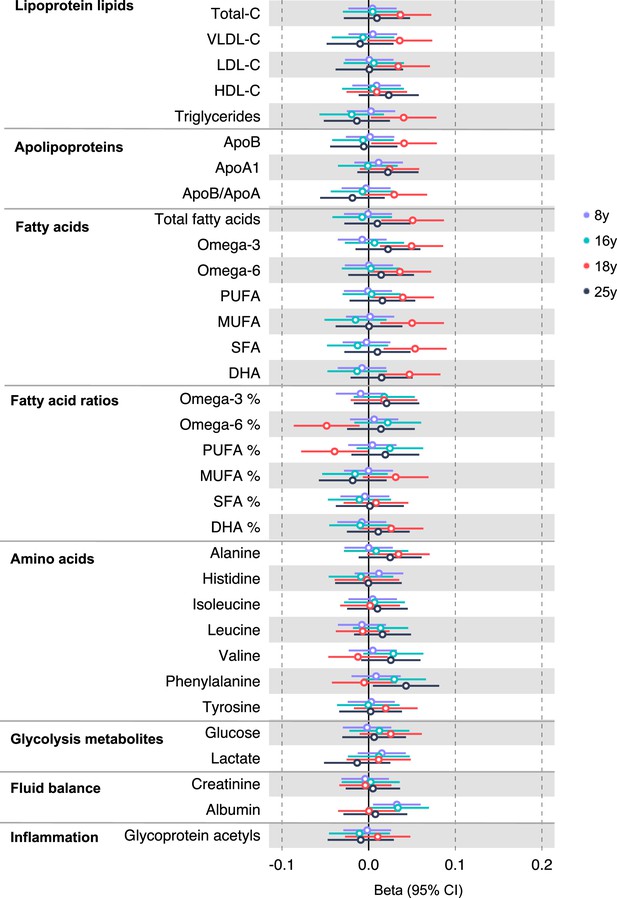
Associations of genetic liability to adult colon cancer (excluding rs174535) with clinically validated metabolic traits at different early life stages among the Avon Longitudinal Study of Parents and Children (ALSPAC) offspring (age 8 y, 16 y, 18 y, and 25 y).
Estimates shown are beta coefficients representing the SD difference in metabolic trait per doubling of genetic liability to colorectal cancer (purple, 8 y; turquoise, 16 y; red, 18 y; black, 25 y). Filled point estimates are those that pass a Benjamini–Hochberg FDR multiple-testing correction (FDR < 0.05).
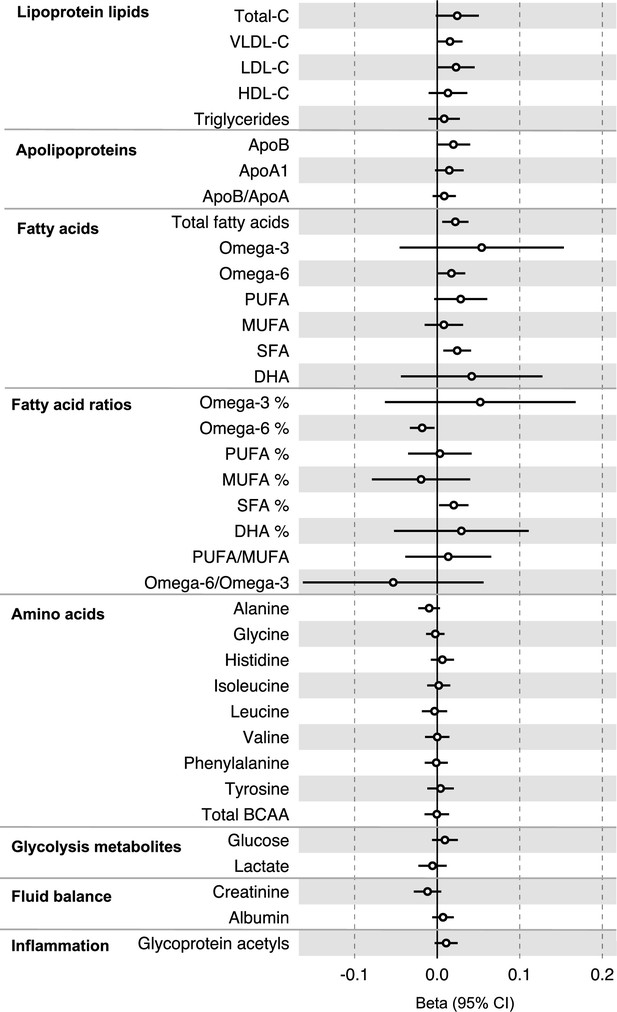
Associations of genetic liability to colorectal cancer with clinically validated metabolic traits in an independent sample of adults (UK Biobank, N = 118,466, median age 58 y) based on reverse two-sample Mendelian randomization analyses.
Estimates shown are beta coefficients representing the SD-unit difference in metabolic trait per doubling of liability to colorectal cancer. Filled point estimates are those that pass a Benjamini–Hochberg FDR multiple-testing correction (FDR < 0.05).
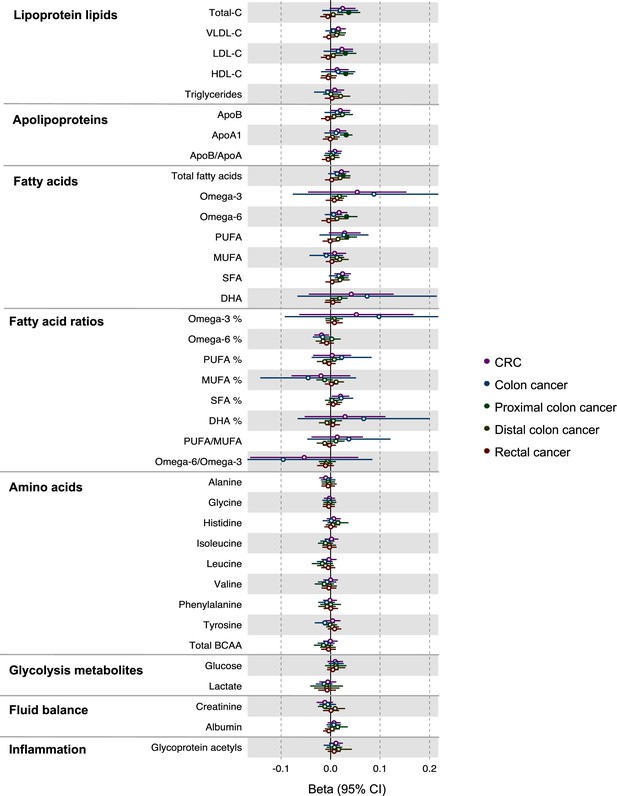
Associations of genetic liability to colorectal cancer with clinically validated metabolic traits in an independent sample of adults based on reverse two-sample Mendelian randomization analyses.
Estimates shown are beta coefficients representing the SD-unit difference in metabolic trait per doubling of liability to colorectal cancer by site (colorectal, colon, distal colon, proximal colon, and rectal cancer). Filled point estimates are those that pass a Benjamini–Hochberg FDR multiple-testing correction (FDR < 0.05).

Associations of genetic liability to colorectal cancer (excluding genetic variants in the FADS gene region) with clinically validated metabolic traits in an independent sample of adults based on reverse two-sample Mendelian randomization analyses.
Estimates shown are beta coefficients representing the SD-unit difference in metabolic trait per doubling of liability to colorectal cancer. Filled point estimates are those that pass a Benjamini–Hochberg FDR multiple-testing correction (FDR < 0.05).
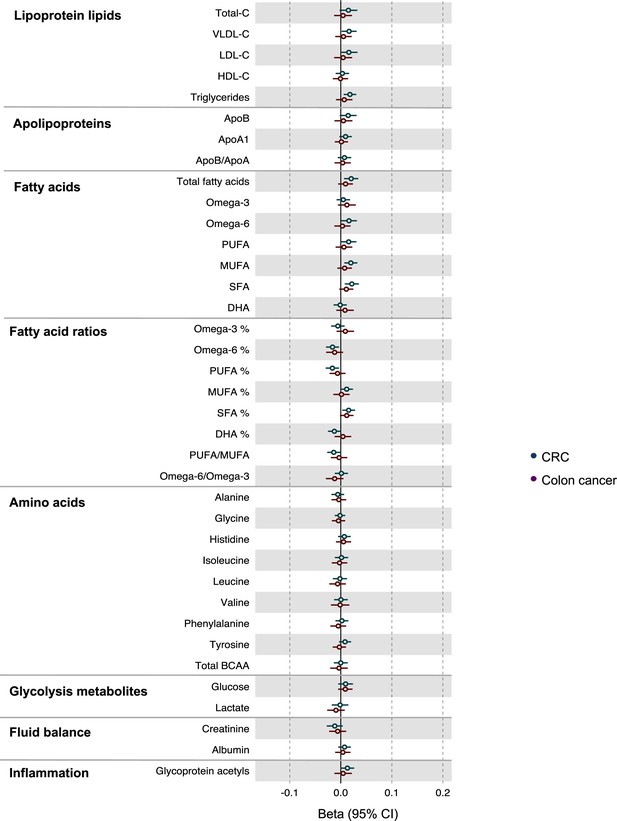
Associations of genetic liability to colorectal and colon cancer with clinically validated metabolic traits in an independent sample of adults based on reverse two-sample Mendelian randomization analyses with FADS variants excluded from colorectal cancer instruments.
Estimates shown are beta coefficients representing the SD-unit difference in metabolic trait per doubling of liability to colorectal cancer by site (colorectal, colon). Filled point estimates are those that pass a Benjamini–Hochberg FDR multiple-testing correction (FDR < 0.05).
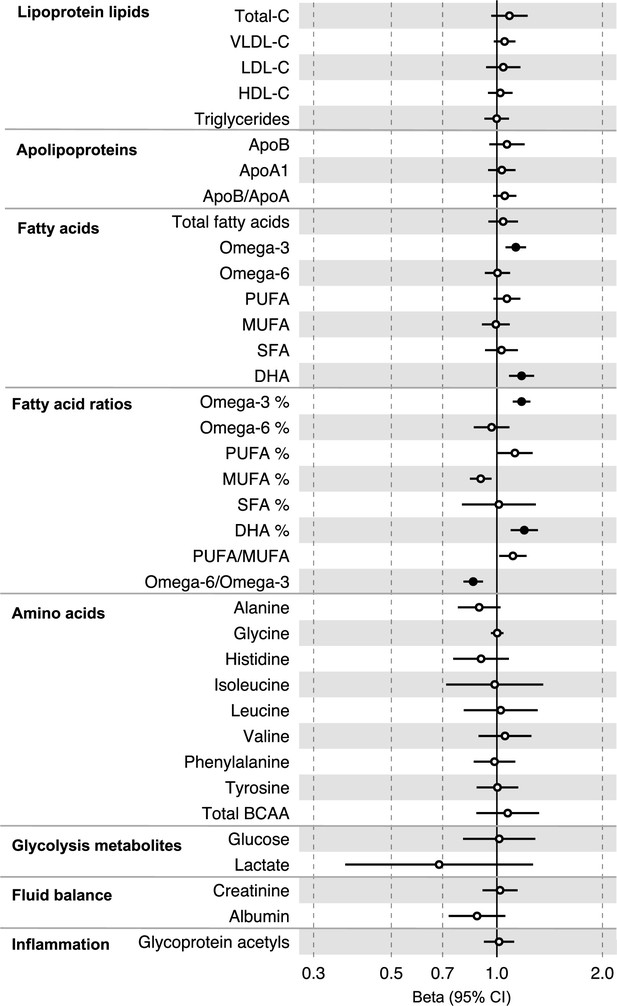
Associations of clinically validated metabolites with colorectal cancer based on conventional (forward) two-sample Mendelian randomization analyses in individuals from UK Biobank (N = 118,466, median age 58 y).
Estimates shown are beta coefficients representing the logOR for colorectal cancer per SD metabolite. Filled point estimates are those that pass a Benjamini–Hochberg FDR multiple-testing correction (FDR < 0.05).
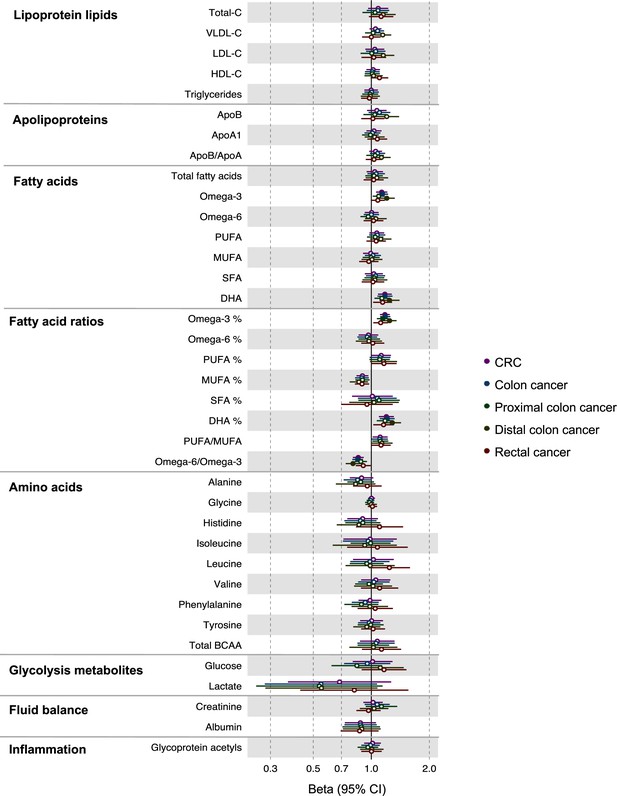
Associations of clinically validated metabolites with colorectal cancer by site (colorectal, colon, distal colon, proximal colon, and rectal cancer) based on conventional (forward) two-sample Mendelian randomization analyses.
Estimates shown are ORs for colorectal cancer per SD metabolite. Filled point estimates are those that pass a Benjamini–Hochberg FDR multiple-testing correction (FDR < 0.05).
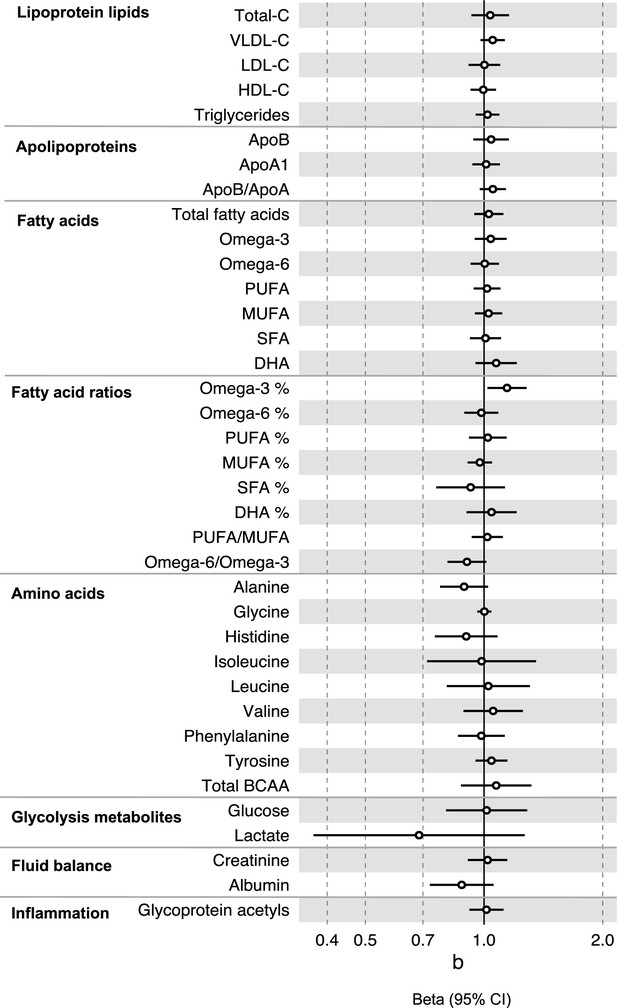
Associations of clinically validated metabolites with colorectal cancer based on conventional (forward) two sample Mendelian randomization analyses with FADS variants excluded from metabolite instruments.
Estimates shown are ORs for colorectal cancer per SD metabolite. Filled point estimates are those that pass a Benjamini–Hochberg FDR multiple-testing correction (FDR < 0.05).
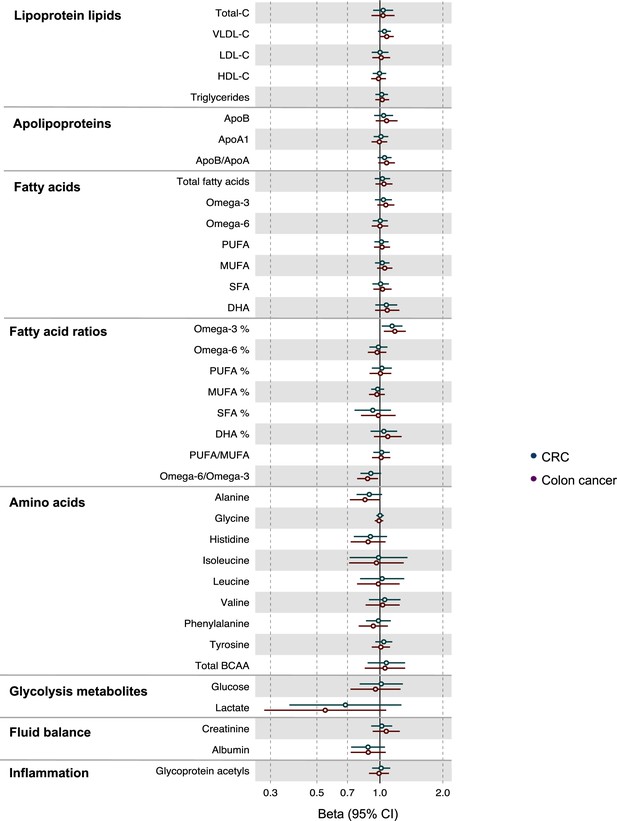
Associations of clinically validated metabolites with colorectal cancer by site (colorectal, colon) based on conventional (forward) two sample Mendelian randomization analyses with FADS variants excluded from metabolite instruments.
Estimates shown are ORs for colorectal cancer per SD metabolite. Filled point estimates are those that pass a Benjamini–Hochberg FDR multiple-testing correction (FDR < 0.05).
Additional files
-
Supplementary file 1
Supplementary tables.
(a) Genetic variants used to construct genetic risk scores reflecting colorectal cancer liability. (b) Mean and SD values for raw metabolic traits at different life stages among ALSPAC offspring. (c) Associations of genetic liability to colorectal cancer with metabolic traits at different early life stages among ALSPAC offspring. (d) Associations of genetic liability to colorectal cancer (excluding SNPs in the FADS gene region) with metabolic traits at different early life stages among ALSPAC offspring. (e) Assessment of instrument strength for MR analyses. (f) Associations of genetic liability to colorectal cancer with metabolic traits based on two-sample MR. (g) Associations of genetic liability to colorectal cancer with metabolic traits based on two-sample MR, excluding variants in the FADS gene region. (h) Genetic variants used to instrument circulating metabolites. (i) Assessment of instrument strength for MR analyses. (j) Estimated effects of circulating metabolites on colorectal cancer risk based on two-sample MR. (k) Estimated effects of circulating metabolites on colorectal cancer risk based on two-sample MR, excluding variants in the FADS gene region.
- https://cdn.elifesciences.org/articles/87894/elife-87894-supp1-v2.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/87894/elife-87894-mdarchecklist1-v2.docx





