Sensitive remote homology search by local alignment of small positional embeddings from protein language models
Figures
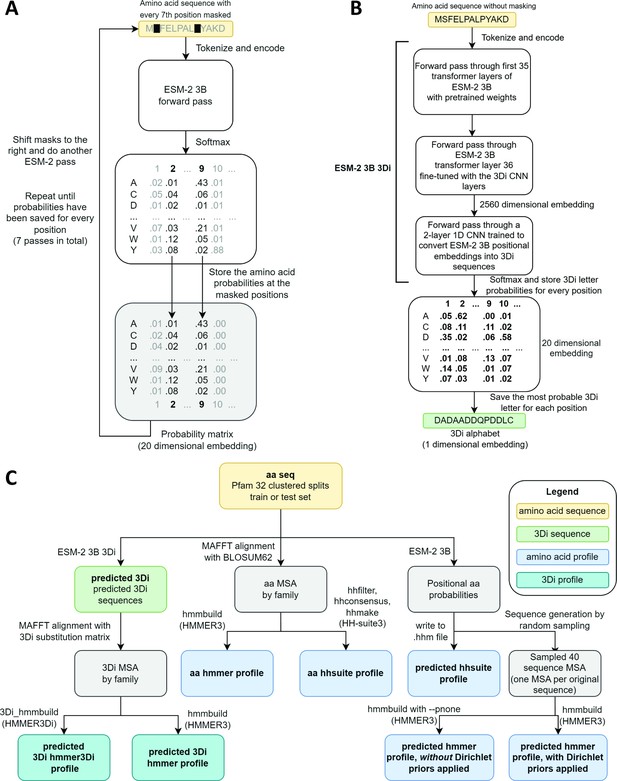
Schematics of embedding models and the experimental design.
(A) ESM-2 3B can be directly used to predict amino acid probability distributions at masked positions. Our implementation uses seven passes. The second pass is shown in the figure. (B) ESM-2 3B 3Di, a fine-tuned ESM-2 3B with a small convolutional neural network (CNN) top model can be used to predict 3D interaction (3Di) sequences from amino acid sequences. (C) Data flow from amino acid sequences through embedding models and other programs to produce files used in homology searches. Bold words correspond to line labels in Figure 3.
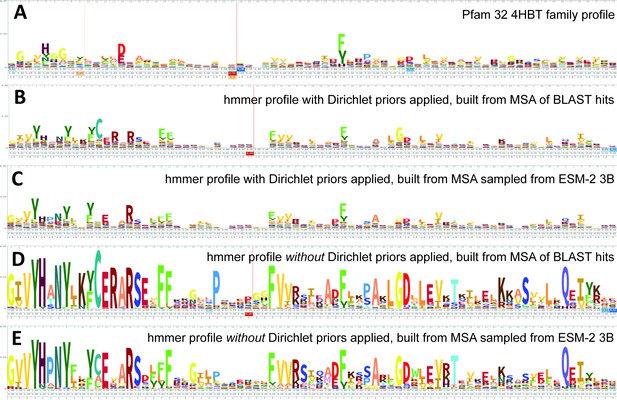
Logos related to the example test sequence YBGC_HELPY___14–90 from the 4HBT family.
(A) 4HBT family hmm from Pfam 32. (B) hmmbuild with default settings on a multiple sequence alignment (MSA) of the top 100 hits supplied by an online blast search (https://blast.ncbi.nlm.nih.gov/Blast.cgi) of YBGC_HELPY___14–90 against the NCBI clustered nr database. (C) hmmbuild with default settings on the MSA sampled from the ESM-2 3B positional probabilities for YBGC_HELPY___14–90. (D, E) hmmbuild with Dirichlet priors disabled on the same MSAs as for (B, C), respectively. All logos were generated by uploading the corresponding.hmm file to https://skylign.org/ (Wheeler et al., 2014).

Homology detection accuracy.
Test sequences were binned based on percent identity to the closest training sequence in the same family and annotated based on the top scoring hit from a search against the entire set of training sequences or training sequence family profiles, depending on the algorithm. (A, C) Family recovery accuracy by bin. (B, D) Clan recovery accuracy. (A, B) Compare amino acid profile-based methods. (C, D) Compare Foldseek-based methods. Dashed lines are controls. There are a total of 21,293 test sequences. 12,246 test sequences have clan assignments.
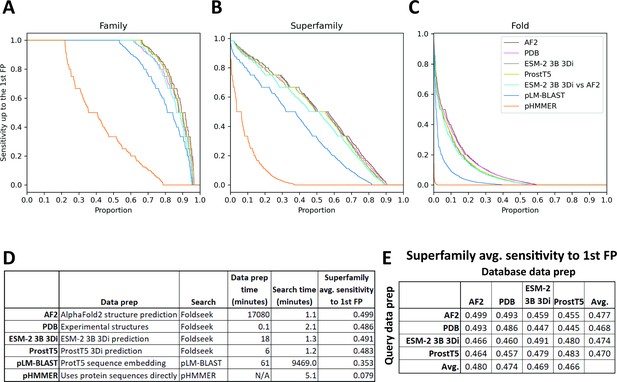
SCOPe40 benchmark.
Cumulative distribution plots of the number of queries attaining each level of sensitivity to the first false positive fold at the (A) family, (B) superfamily, and (C) fold level. (D) Data preparation, search times, and average sensitivity at the superfamily level. (E) Comparison of average sensitivity at the superfamily level of Foldseek run with queries and databases prepared by different methods.





