Recording and classifying MET receptor mutations in cancers
Figures
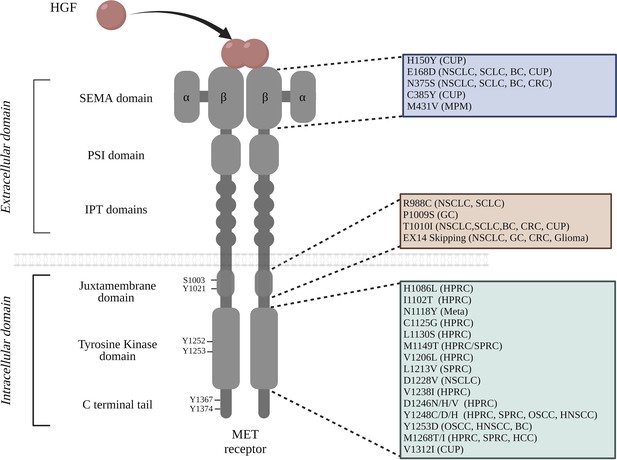
List of MET mutations found within the functional domains of the receptor.
The extracellular portion of MET consists of the SEMA domain, a PSI domain, and four immunoglobulin-plexin-transcription (IPT) repeats; the intracellular region contains the juxtamembrane domain, the tyrosine kinase domain, and the carboxyterminal docking site. The cancer types in which particular mutations have been identified are noted in parentheses: breast cancer (BC), cancer of unknown primary origin (CUP), colorectal cancer (CRC), gastric cancer (GC), hepatocellular carcinoma (HCC), hereditary papillary renal carcinoma (HPRC), non-small cell lung cancer (NSCLC), small cell lung cancer (SCLC), sporadic papillary renal carcinoma (SPRC), malignant pleural mesothelioma (MPM), oropharyngeal squamous cell carcinoma (OSCC), and metastasis (Meta). It is worth noting that the amino acid positions are annotated from MET transcript variant 1 NM_001127500.3 (Tovar and Graveel, 2017; Ma et al., 2005; Ma et al., 2003; Liu et al., 2015; Jardim et al., 2014; Stella et al., 2011; Jagadeeswaran et al., 2006; Moon et al., 2000; Park et al., 1999Bahcall et al., 2016).
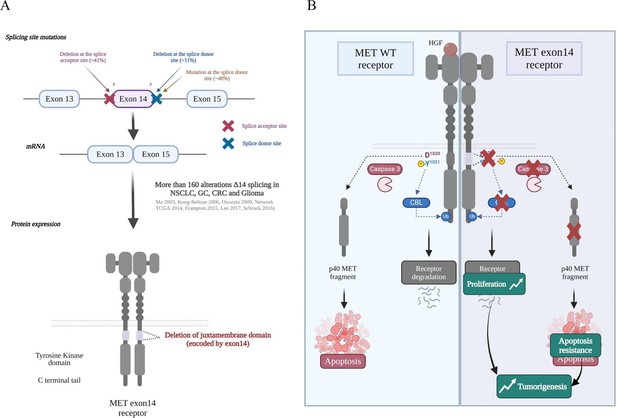
MET mutations leading to exon 14 skipping.
(A) Several mutations affecting the splice junctions flanking exon 14 (encoding the juxtamembrane domain) have been described in non-small cell lung cancer (NSCLC). More than 160 alterations of the exon 14 splicing sites have been described to induce exon skipping and deletion of the MET juxtamembrane domain (Tovar and Graveel, 2017; Ma et al., 2003; Frampton et al., 2015; Lee et al., 2017). (B) Under physiological conditions, MET is also phosphorylated on Tyr1021 in the justamembrane domain, which induces recruitment of Casitas B-lineage lymphoma (CBL), an E3 ubiquitin ligase involved in ubiquitination of the receptor. The ubiquitinated receptor is internalized and degraded, thereby contributing to attenuated signalling. In the absence of its ligand and in response to apoptotic stress, MET is cleaved by caspases to a fragment (p40 MET) able to amplify cell death through permeabilization of the mitochondria. Deletion of the juxtamembrane domain encoded by exon 14 can potentially lead to increased MET stability and tyrosine kinase activity and resistance to cell death. The subsequent increase in proliferation, motility, migration, invasion, and survival promotes tumorigenesis.

Resistance mutations in the METex14Del variant after treatment with a MET TKI.
(A) Upon treatment of MET exon 14 skipping NSCLC patients with a type I MET TKI (able to bind the active form of MET kinase) or a type II MET TKI (able to bind the inactive form of MET kinase), secondary resistance mutations were identified. Some of them (*) were already known as activating mutations in papillary renal cancer. Note that the amino acid positions are annotated from MET transcript variant NM_000245.3 (Schmidt et al., 1997; Tovar and Graveel, 2017). (B) Secondary activating MET mutations were placed on the 3D structure of the MET kinase domain, along with crizotinib (https://doi.org/10.2210/pdb2WGJ/pdb) or merestinib (https://doi.org/10.2210/pdb4EEV/pdb). MET mutations were inserted into the raw PDB data for MET (Research Collaboratory for Structural Bioinformatics Protein Data Bank DOI: 10.2210/pdb2WGJ/pdb and 10.2210/pdb4EEV/pdb).
Resistance mutations of METex14Del treated with MET TKI crizotinib.
Video of the three-dimensional crystal structure of the MET kinase domain, along with crizotinib (https://doi.org/10.2210/pdb2WGJ/pdb). Crizotinib (TKI type I) binds the active state (DFG sequence -in) of the kinase. MET resistance mutations were inserted into the raw PDB data for MET (Research Collaboratory for Structural Bioinformatics Protein Data Bank DOI: 10.2210/pdb2WGJ/pdb). The figures and spin structure videos were created with PyMOL software and extracted. The colour code for mutations is the same as in Figure 3.
Resistance mutations of METex14Del treated with MET TKI meristinib.
Video of the three-dimensional crystal structure of the MET kinase domain, along with merestinib (https://doi.org/10.2210/pdb4EEV/pdb). Merestinib (TKI type II) binds the inactive stage (DFG-out). MET resistance mutations were inserted into the raw PDB data for MET (Research Collaboratory for Structural Bioinformatics Protein Data Bank DOI: 10.2210/pdb4EEV/pdb). The figures and spin structure videos were created with PyMOL software and extracted. The colour code for mutations is the same as in Figure 3.
Tables
Recording of MET mutations found in cancers (Tovar and Graveel, 2017; Ma et al., 2005; Ma et al., 2003; Liu et al., 2015; Jardim et al., 2014; Stella et al., 2011; Jagadeeswaran et al., 2006; Moon et al., 2000; Park et al., 1999; Bahcall et al., 2016).
In this table, recording the MET mutations characterized by functional studies, subdomain localization of MET mutations were indicated as well as their sensitivity to hepatocyte growth factor (HGF) stimulation and the cancer type in which they were identified. The type of functional assay performed to identify them as activating mutations is indicated.
| Domain | Subdomain | AA | Cancer | Functional assay | Sensitivity to HGF | References |
|---|---|---|---|---|---|---|
| Extracellular | SEMA | H150Y | CUP | Anchorage-independent growth assay | Stella et al., 2011 | |
| E168D | NSCLC, SCLC, BC, CUP | BaF3 cells, soft agar colony assay SCLC H446 | Ma et al., 2003 | |||
| N375S | NSCLC, SCLC, BC, CRC | Cell migration, invasion, and colony-forming assay, tumor growth study after cell xenograft | Ma et al., 2005; Jardim et al., 2014 | |||
| C385Y | CUP | Anchorage-independent growth assay | Stella et al., 2011 | |||
| M431V | Malignant pleural mesothelioma (MPM) | Cell migration and motility assay | Jagadeeswaran et al., 2006 | |||
| Juxtamembrane | R988C | NSCLC, SCLC | BaF3 cells, soft agar colony assay SCLC H446 | Ma et al., 2003; Montagne et al., 2017 | ||
| P1009S | GC | Focus formation NIH3T3 | Lee et al., 2000 | |||
| T1010I | NSCLC, SCLC, BC, CRC, CUP | Focus formation NIH3T3 | Lee et al., 2000 | |||
| Kinase N-lobe | H1086L | HPRC | Focus formation NIH3T3 | Yes | Sebai et al., 2022 | |
| P-Loop | I1102T | HPRC | Focus formation NIH3T3 | Yes | Sebai et al., 2022 | |
| P-Loop | N1118Y | Metastasis | Cell migration and invasion assay | Yes | Lorenzato et al., 2002 | |
| P-Loop | C1125G | HPRC | Focus formation NIH3T3 | Yes | Sebai et al., 2022 | |
| P-Loop | L1130S | HPRC | Focus formation NIH3T3 | Yes | Sebai et al., 2022 | |
| M1149T | HPRC/SPRC | Focus formation NIH3T3 | Yes | Jeffers et al., 1997; Michieli et al., 1999 | ||
| Kinase C-lobe | V1206L | HPRC | Focus formation NIH3T3 | Yes | Jeffers et al., 1997; Michieli et al., 1999 | |
| L1213V | SPRC | Focus formation NIH3T3 | Yes | Michieli et al., 1999 | ||
| Activation Loop (1231–1262) | V1238I | HPRC | Focus formation NIH3T3 | Yes | Jeffers et al., 1997; Michieli et al., 1999 | |
| Activation Loop (1231–1262) | D1246N/H/V | HPRC | Focus formation NIH3T3 | Yes | Jeffers et al., 1997; Michieli et al., 1999; Bahcall et al., 2016 | |
| Activation Loop (1231–1262) | Y1248C/D/H | HPRC, SPRC, OSCC, HNSCC | Focus formation NIH3T3 | Yes | Jeffers et al., 1997; Michieli et al., 1999 | |
| Activation Loop (1231–1262) | Y1253D | OSCC, HNSCC, BC | Focus formation NIH3T3 | Jeffers et al., 1997; Bardelli et al., 1998; Liu et al., 2015 | ||
| COOH-terminal lobe of the kinase domain | M1268T/I | HPRC, SPRC, HCC | Focus formation NIH3T3 | Jeffers et al., 1997; Michieli et al., 1999 | ||
| V1312I | CUP | Anchorage-independent growth assay | Stella et al., 2011 |





