Role of Tim17 in coupling the import motor to the translocation channel of the mitochondrial presequence translocase
Figures
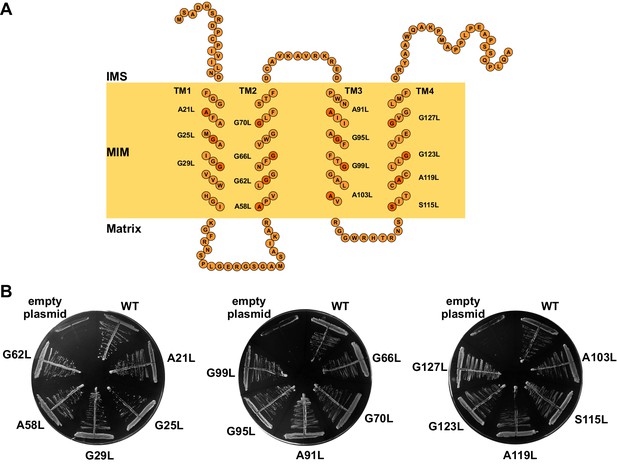
The GxxxG motifs in transmembrane segments of Tim17.
(A) Schematic representation of predicted topology of Tim17. Glycine, alanine or serine residues in GxxxG and GxxxG-like motifs are highlighted. IMS, intermembrane space. MIM, mitochondrial inner membrane. (B) Each residue of the motifs was replaced by leucine. The ability of obtained mutants to complement TIM17 deletion was analyzed on a medium containing 5-FOA. Empty plasmid and a plasmid encoding a wt version of Tim17 were used as negative and positive controls, respectively.
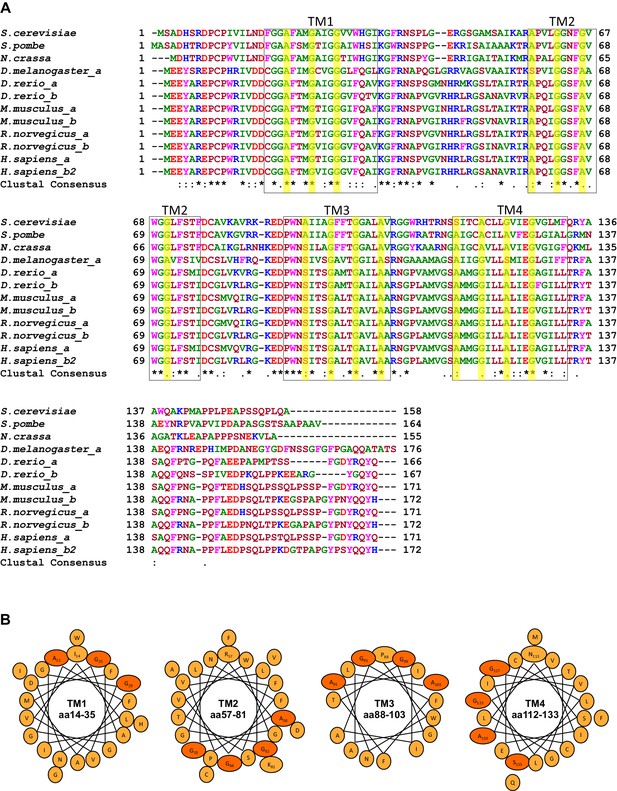
Conserved GxxxG motifs in the transmembrane segments of Tim17.
(A) Multiple sequence alignment of Tim17 proteins from various species. Saccharomyces cerevisiae, Schizosaccharomyces pombe, Neurospora crassa, Drosophila melanogaster, Danio rerio, Mus musculus, Rattus norvegicus, Homo sapiens. Predicted transmembrane (TM) segments are in boxes. The glycine, alanine and serine residues of GxxxG and GxxxG-like motifs are highlighted. (B) Helical wheel projection of TMs 1-4 of Tim17. The glycine, alanine and serine residues of GxxxG and GxxxG-like motifs residues are shown in orange.
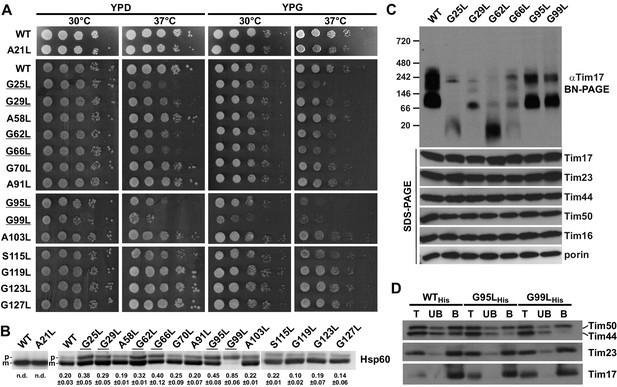
Effects of mutations in GxxxG motifs.
(A) Ten-fold serial dilutions of cells were spotted on YPD and YPG plates and incubated at 30°C and 37°C. Temperature-sensitive mutants are underlined. (B) Total cell extracts of cells grown at 37°C were analyzed by SDS-PAGE and immunoblotting with Hsp60 antibodies. p, precursor and m, mature forms of Hsp60. Fraction of precursor form compared to total signal (precursor plus mature form) was quantified from three experiments and shown as mean with standard deviation. n.d. – none detectable (in three experiments). Temperature-sensitive mutants are underlined. (C) Isolated mitochondria, as indicated, were solubilized with 1% digitonin and analyzed by Blue Native- (upper panel) and SDS-PAGE (lower panel) and immunoblotting with indicated antibodies. (D) Mitochondria were isolated from cells expressing indicated Tim17 versions with a C-terminal His-tag, solubilized in digitonin-containing buffer and cleared lysates were incubated with Ni-agarose beads. After three washing steps, specifically bound material was eluted with Laemmli buffer containing 300 mM imidazole. Total (T, 20%), unbound (UB, 20%) and bound (B, 100%) fractions were analyzed by SDS-PAGE and immunoblotting with indicated antibodies.
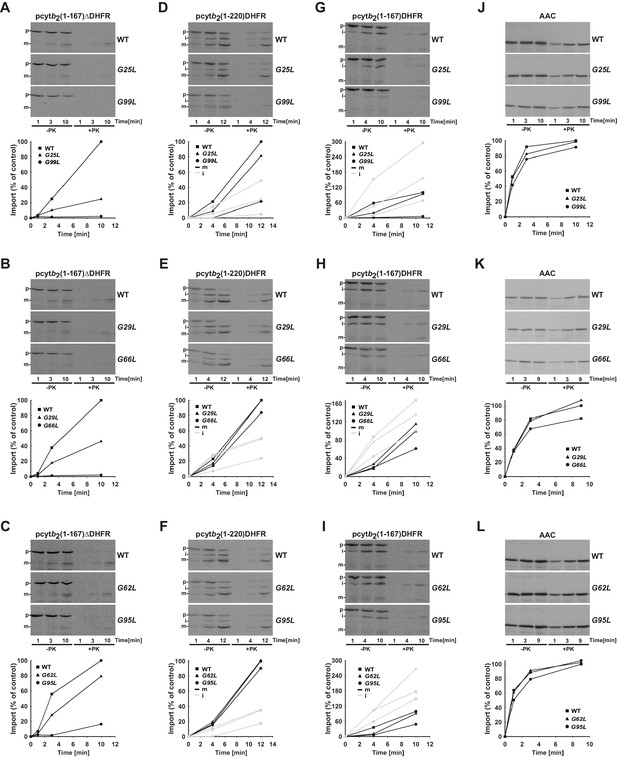
Temperature-sensitive mutations in Tim17 affect in vitro import of proteins by the TIM23 complex.
(A–L) Wild type (WT) and indicated ts mutant yeast cells were grown under permissive conditions in YPD medium at 24°C and mitochondria were subsequently isolated. TIM23-dependent mitochondrial precursor proteins, a matrix targeted pcytb2(1–167)△DHFR and laterally sorted pcytb2(1–167)DHFR and pcytb2(1–220)DHFR were synthesized in vitro in the presence of 35S-methionine and incubated with isolated mitochondria that were preincubated for 30 min at 37°C. At indicated time points, samples were taken out and diluted into ice-cold buffer containing valinomycin to stop further import. One set of samples was treated with Proteinase K (PK) and the other was left untreated. All samples were analyzed by SDS-PAGE followed by transfer onto nitrocellulose membrane and autoradiography (upper panels). In the lower panels, signals after PK-treatment were quantified using Image J software. The intensity of the mature band in the longest time point of import into WT mitochondria was set to 100%. Essentially the same experiment was done with a TIM23-independent precursor, ATP/ADP carrier (AAC), as a control. p - precursor, i - intermediate and m - mature forms of imported precursors.
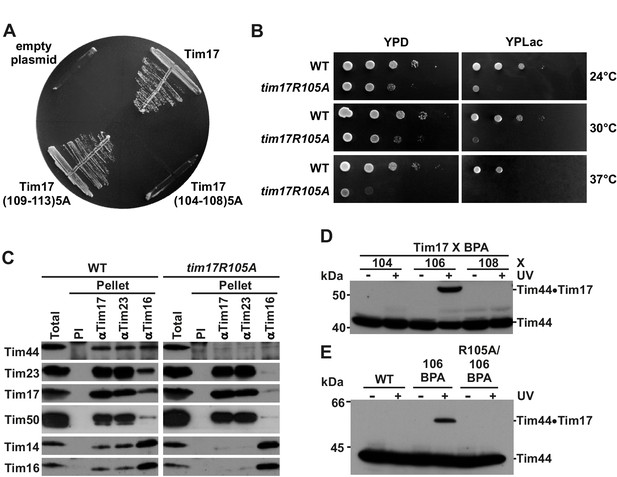
Identification of a region of Tim17 involved in binding of Tim44.
(A) Alanine-scanning mutagenesis of Tim17 region encompassing residues 104 to 113. Five residues in a row were mutated to alanines and the ability of the mutants to complement TIM17 deletion was analyzed on a medium containing 5-FOA. Empty plasmid served as a negative control and a plasmid encoding wt Tim17 as a positive control. (B) Ten-fold serial dilutions of cells were spotted on YPD and YPLac plates and incubated at indicated temperatures. (C) Isolated mitochondria were solubilized with digitonin and incubated with affinity purified antibodies to Tim17, Tim23 and Tim16 prebound to Protein A-Sepharose. Antibodies from preimmune serum (PI) were used as a negative control. Total (15%) and material specifically bound to the beads (100%) were analyzed by SDS-PAGE and immunoblotting with the indicated antibodies. (D) p-benzoylphenylalanine (BPA) was introduced at positions 104, 106 and 108 of Tim17. Where indicated, cells were UV-irradiated. Total cell extracts were prepared and analyzed by SDS-PAGE and immunoblotting with antibodies to Tim44. (E) Indicated cells were treated and analyzed as in panel D.
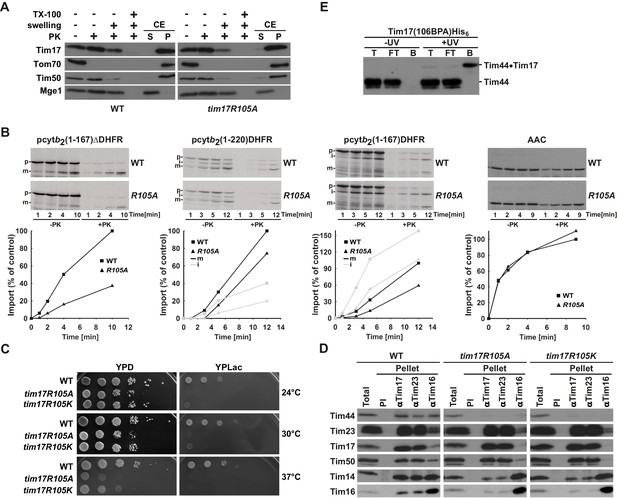
Region of Tim17 involved in binding of Tim44.
(A) Tim17R105A mutant is correctly inserted into the inner membrane. Submitochondrial localization of Tim17, wild type (WT) and R105A mutant (tim17R105A), was analyzed by its accessibility to Proteinase K (PK) in intact mitochondria, upon opening of the outer membrane (+ swelling) and upon solubilization of both mitochondrial membranes (+TX-100). In addition, mitochondria were subjected to carbonate extraction (CE) and fractionated into supernatant (S) and pellet (P) fractions that contain soluble and membrane proteins, respectively. All samples were analyzed by SDS-PAGE followed by immunoblotting with indicated antibodies. Tom70 served as a marker for a membrane-integrated outer membrane protein, Tim50 for a membrane-integrated inner membrane protein exposed to the intermembrane space and Mge1 for a soluble matrix protein. (B) Mitochondria were isolated from wild type (WT) and tim17R105A (R105A) yeast cells grown under permissive conditions in YPD medium at 24°C. In vitro imports of indicated precursor proteins were performed as described in Figure 2—figure supplement 1. (C) Ten-fold serial dilutions of cells were spotted on YPD and YPLac plates and incubated at indicated temperatures. (D) Isolated mitochondria, as indicated, were solubilized in digitonin-containing buffer and cleared lysates were incubated with affinity purified antibodies to Tim17, Tim23 and Tim16 prebound to Protein A-Sepharose. Antibodies from preimmune (PI) serum were used as a negative control. After three washing steps, proteins specifically bound to the beads were eluted with Laemmli buffer. Total (20%) and bound fractions (100%) were analyzed by SDS-PAGE followed by immunoblotting with indicated antibodies. (E) Cells carrying a His-tagged version of Tim17 and a BPA residue at position 106 were UV irradiated, where indicated. Total cell extracts were prepared, diluted with Triton X-100 buffer and incubated with Ni-agarose beads. After three washing steps, specifically bound proteins were eluted. Samples were analyzed by SDS-PAGE and immunoblotting with Tim44 antibodies.
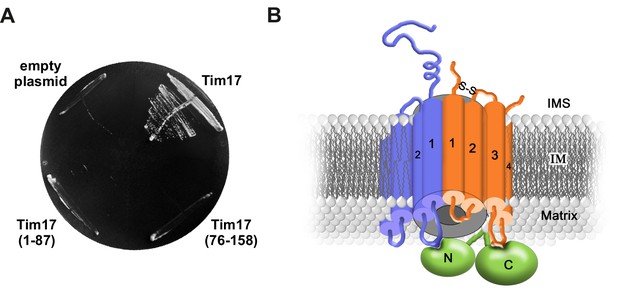
Transmembrane segments of Tim17 have functionally distinct roles.
(A) The ability of the indicated mutants to complement TIM17 deletion was analyzed on a medium containing 5-FOA. (B) A model of Tim17 (orange) and its interactions with Tim23 (blue) and Tim44 (green). See text for details.
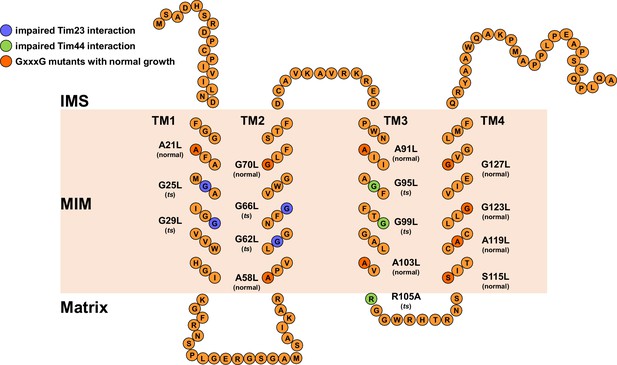
Schematic representation of Tim17 and its interactions.
Residues that were mutated in this study are highlighted. Residues whose mutations affected interaction of Tim17 with Tim23 are shown in blue and the ones that affected interaction with Tim44 in green. Mutations that were made but had no obvious growth defect are shown in dark orange. IMS, intermembrane space. MIM, mitochondrial inner membrane.





