Mutation analysis links angioimmunoblastic T-cell lymphoma to clonal hematopoiesis and smoking
Figures
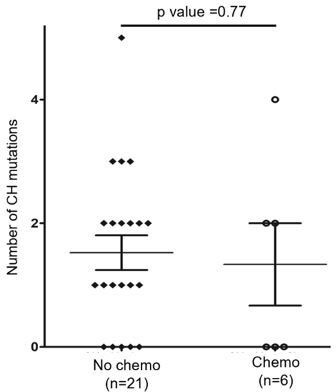
Analysis of genomic alterations by target sequencing panel for primary lymphomas and paired bone marrow/peripheral blood (BM/PB) in patients with angioimmunoblastic T-cell lymphoma (AITL) and peripheral T-cell lymphoma, not otherwise specified (PTCL-NOS).
(A) Presence of clonal hematopoiesis (CH) in patients with AITL and PTCL-NOS. Dot plots showing the detected variants and their variant allele frequencies (VAFs) in the AITL and PTCL-NOS (lymph node [LN]) and their matched BM/PB in representative AITL and PTCL-NOS cases with CH. The black circles indicate variants specific to the lymphomas, and the variants shared between the primary lymphomas and CH are highlighted in red. The variants attributed to lymphoma only are boxed. Additional detailed descriptions of these illustrative cases are provided in Appendix 1. (B) Venn diagram showing the distribution of the shared, lymphoma or BM/PB-specific variants identified in the diagnostic LN and paired BM/PB samples. The shared variants are defined as variants identified in both the primary lymphoma and the BM/PB, the latter as CH-related variants. The variants predicted to be due only to lymphoma involvement in BM/PB have been excluded (see also Figure 1—figure supplement 1A for the distribution of all variants). (C) Summary of the CH-associated mutations in the BM/PB and LN, and the mutations postulated to accumulate at a later stage of lymphoma development (late mutations). The CH-associated mutations are shared between the primary lymphomas and the BM/PB and can be considered as early lesions in AITL/PTCL. The heatmaps show the top recurrent mutations in both categories. Stacked bar plots show the type of variants and the mutation frequency (relative to our cohort) for each of the major mutated genes in the LN and BM/PB samples. Pt: patient; T: tumor. (D) Comparison of the distribution of disruptive and missense mutations in the CH-associated and late mutations. Statistical significance was determined by (D) z test measuring proportion difference. *p<0.05; **p<0.01; ***p<0.001; ****p,0.0001; NS, not significant. P-Value<0.05 is considered statistically significant.
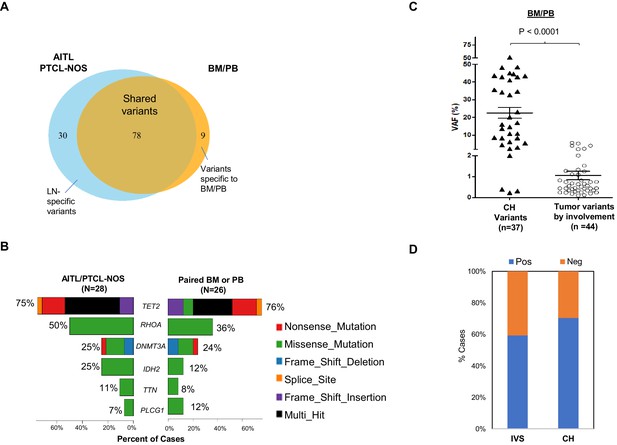
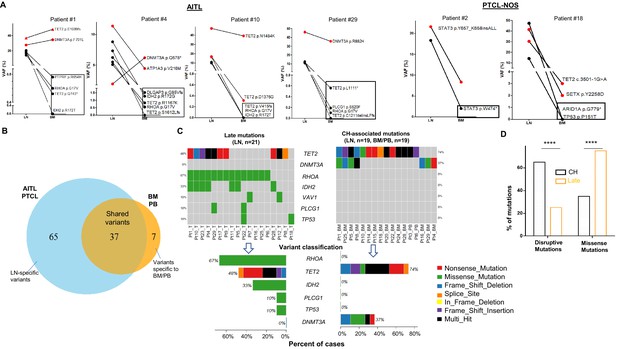
Angioimmunoblastic T-cell lymphoma (AITL) variants involving bone marrow/peripheral blood (BM/PB).
(A) Venn diagram illustrates the distribution of the shared, and the lymph node (LN) or BM/PB-specific mutations. Note that the variants detected in the BM/PB due to lymphoma involvement are included in this diagram, in contrast to Figure 1B presented in the main text. (B) The type of variants and mutation frequencies (relative to the total number of cases) for the top six mutated genes identified in AITL/peripheral T-cell lymphoma, not otherwise specified (PTCL-NOS) and matched BM/PB are shown. Different colors represent variant classifications as indicated. (C) Dot plot comparing variant allele frequencies (VAFs) of the clonal hematopoiesis (CH)-associated variants and those related to lymphoma involvement in the BM/PB specimens. Mean ± SEM of the VAFs is shown for each subgroup. (D) The proportion of the cases with or without BM or PB involvement by the neoplastic T-cells (IVS), or with/without CH, respectively, is summarized. Statistical significance in (C) was determined by t test. p-Value<0.05 is considered statistically significant.
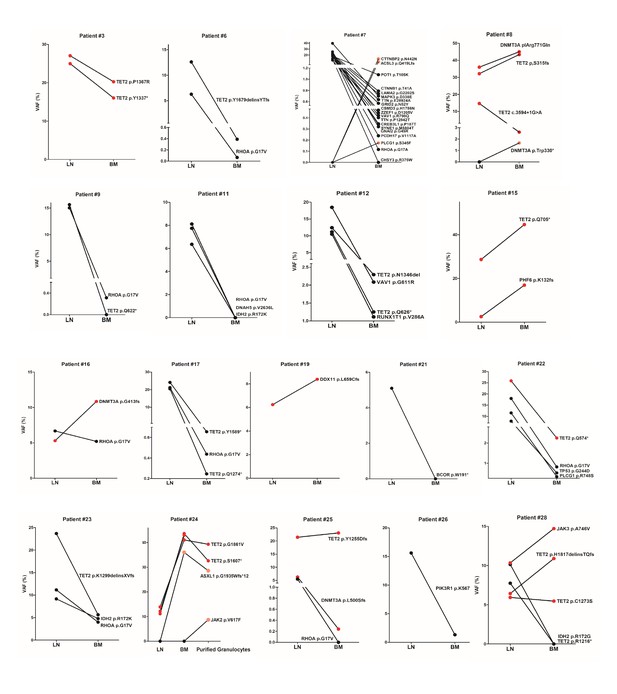
Comparison of variant allele frequencies (VAFs) of the variants found in paired angioimmunoblastic T-cell lymphoma (AITL) and bone marrow/peripheral blood (BM/PB) samples.
Red color highlights the clonal hematopoiesis (CH)-associated variants shared between the lymph node (LN) and BM/PB compartments. The black circles indicate variants specific to the lymphomas, the pink circles represent mutations specific to BM/PB. The cases that were not presented in Figures 1 and 3 are shown in this figure supplement. In patient #7, three mutations specific to BM were found in PLCG1, ACSL3, and CTTNBP2. These may represent novel CH-associated mutations. In patient #8, DNMT3A p.W330*, likely representing a minor CH clone, was also found specifically in the BM. In patient #24, an additional ASXL1 frameshift mutation was identified in the BM, which showed hypercellularity with increased granulopoiesis. In this patient, we appeared to capture the acquisition of a subclonal JAK2 V617F mutation during tumor progression, which was identified by the myeloid panel in the purified granulocytes from the PB but not detected in the BM 1 year before.
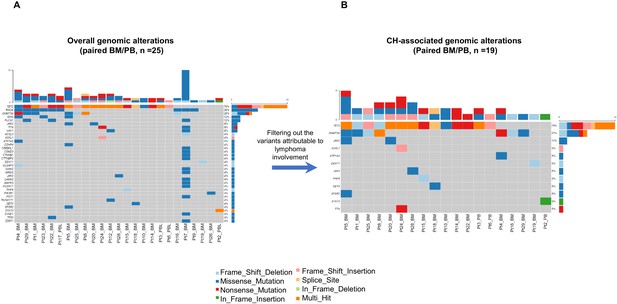
Overall and clonal hematopoiesis (CH)-related genomic alterations in the matched bone marrow/peripheral blood (BM/PB) samples.
(A) The overall mutation profile in the BM/PB is shown. (B) The CH-related mutation profile identified in the BM/PB is shown after excluding the variants due to angioimmunoblastic T-cell lymphoma (AITL) involvement. Top bars on each plot indicate the numbers of the variants detected per sample. The percentage showing gene mutation frequency in the patient cohort is shown at the right. Sample IDs are indicated at the bottom of each plot. Variant classifications are indicated with different colors.
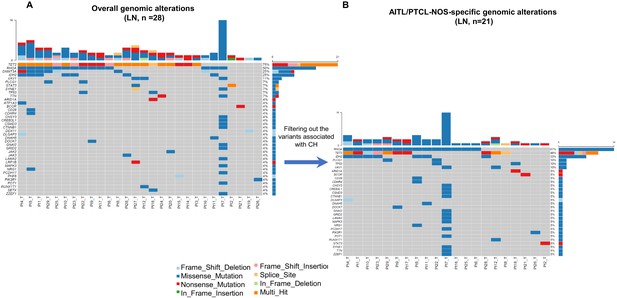
Overall and lymphoma-specific genomic alterations in primary angioimmunoblastic T-cell lymphoma/peripheral T-cell lymphoma, not otherwise specified (AITL/PTCL-NOS).
(A) Mutation plot showing the overall mutation profile in the lymph node (LN) tissues involved by AITL/PTCL-NOS. (B) Mutation plot showing the AITL/PTCL-NOS-specific mutations identified in lymphoma samples after excluding the clonal hematopoiesis (CH)-associated variants. Top bars on each plot indicate the numbers of variants detected per sample. The percentage at right shows the gene mutation frequency in the cohort of the patients. Sample IDs are indicated at the bottom of each plot. Variant classifications are indicated with different colors.
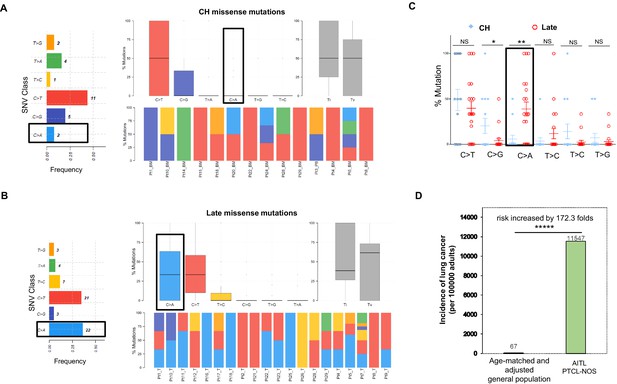
Late mutations in angioimmunoblastic T-cell lymphoma/peripheral T-cell lymphoma, not otherwise specified (AITL/PTCL-NOS) are enriched for C>A transversion substitutions possibly associated with smoking.
Transition and transverse (Titv) plot showing overall distribution of the six types of substitutions in the clonal hematopoiesis (CH) (A) and late (B) missense mutations acquired during AITL/PTCL development, as well as fraction of these substitutions in each sample. The median is indicated by a horizontal line. Bar plot on the left showing single-nucleotide variant (SNV) classes and fraction of each substitution class among all missense mutations. (C) Side-by-side comparison of transition and transversion base substitutions acquired between the early CH-associated and late mutations. (D) Bar plot comparing the incidence rate of lung cancer between two age-matched/-adjusted populations indicated. Statistical significance was determined by (C, D) z test. *p<0.05; **p<0.01; ***p<0.001; ****p<0.00001; NS, not significant.
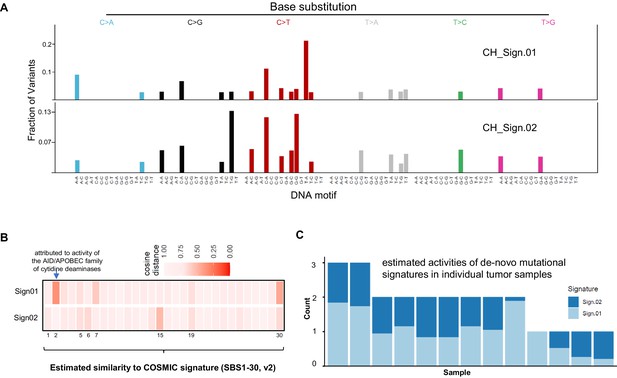
Comparison two de novo mutational signatures from clonal hematopoiesis (CH)-associated angioimmunoblastic T-cell lymphoma (AITL) mutations with COSMIC Signatures.
(A) Profile of two de novo mutational signatures extracted from CH-associated AITL mutations. (B) Heatmap showing similarity of two identified de novo mutation signatures with COSMIC Signature by analysis of cosine distance among these mutation patterns. Numbers on the bottom of the heatmap represent the number of COSMIC Signatures (SBS30, v2). (C) Estimated exposures of each AITL sample to the identified mutational patterns (Sign.01 and Sign.02).
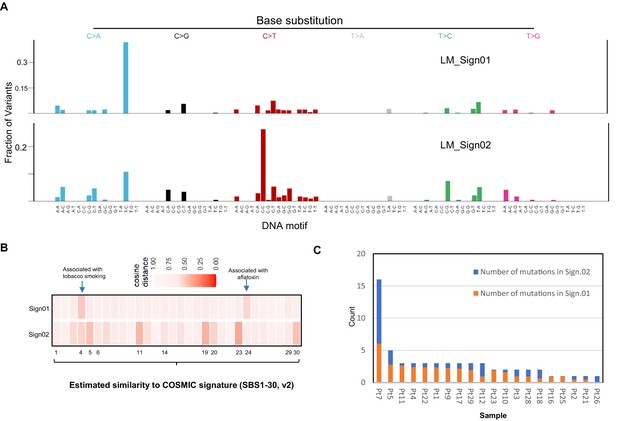
Similarity of two de novo mutational signatures from angioimmunoblastic T-cell lymphoma (AITL) late mutations with COSMIC Signatures.
(A) Profiles of two de novo mutational signatures extracted from AITL late mutations. (B) Heatmap showing similarity of two identified de novo mutation signatures with COSMIC Signature by analysis of cosine similarity. (C) Estimated activities of the identified mutational signatures (LM_Sign01 and LM_Sign02) per sample according to estimated number of individual signature-associated mutations. LM: late mutations; Pt: patient.
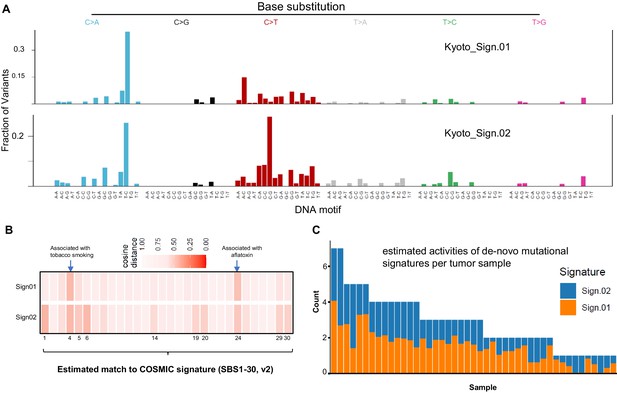
Similarity of two de novo mutational signatures from the published T-follicular helper-peripheral T-cell lymphoma (TFH-PTCL) mutation dataset with COSMIC Signatures.
(A) Profiles of two de novo mutational signatures extracted from published single-nucleotide variant (SNV) mutations from TFH-PTCL tumors (n = 44). (B) Heatmap showing cosine similarity of two identified de novo mutation signatures with 30 COSMIC Signatures. (C) Estimated exposures of each AITL sample to the identified mutational patterns (Kyoto_Sign01 and Kyoto_Sign02).
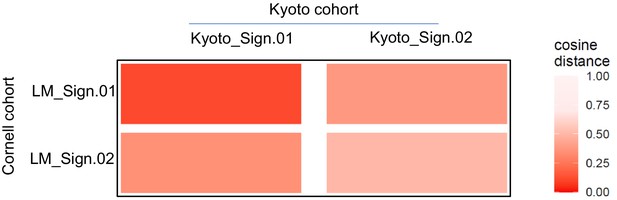
Comparing de novo signatures extracted from angioimmunoblastic T-cell lymphoma (AITL) late mutations and published T-follicular helper-peripheral T-cell lymphoma (TFH-PTCL) mutations.
Heatmap showing cosine similarity of the de novo mutation patterns identified in the indicated mutation data set. LM: AITL late mutations.
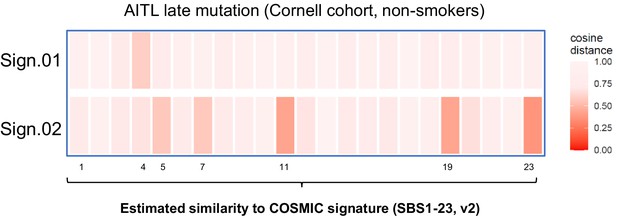
Comparing de novo signatures extracted from angioimmunoblastic T-cell lymphoma (AITL) late mutations in the non-smokers to COSMIC Signatures.
Heatmap showing cosine similarity of the de novo mutation patterns with the COSMIC Signatures.
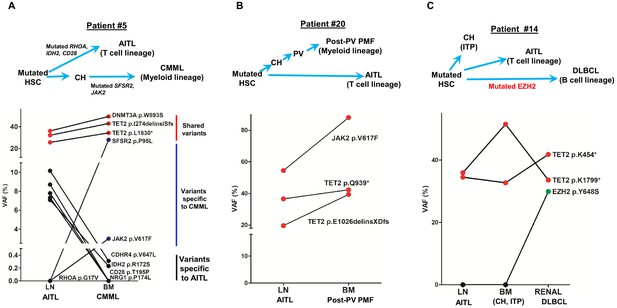
Angioimmunoblastic T-cell lymphoma (AITL) and concomitant hematologic neoplasms develop from common mutated hematopoietic precursor cells.
(A–C) Dot plots comparing variant allele frequencies (VAFs) of the mutations identified in the AITL and the concomitant hematologic malignancies. Red dots show the variants shared between different hematologic neoplasms or entity in the same patient. Dark blue dots in (A) indicate the variants specially related to chronic myelomonocytic leukemia (CMML), and the black dots in (A) denote the AITL-specific variants. Schematic diagrams depicting hypothetical clonal evolution models of the tumors deriving from mutated hematopoietic stem cells (HSC) are also presented. In patient #5 (A), additional mutations besides the clonal hematopoiesis (CH)-associated mutations were identified and implicated in the disease progression to AITL and CMML, respectively. In patient #20, no additional mutations besides those mutated in HSC are identified. In patient #14, a mutated EZH2, indicated by green dot, is implicated in the progression to diffuse large B-cell lymphoma (DLBCL). In all three cases, there are mutations that are shared between the AITL and the concomitant myeloid or B lymphomas, supporting evolution of these neoplasms from a common precursor. PV: polycythemia vena; post-PV PMF, post-PV primary myelofibrosis; ITP: immune thrombocytopenia.
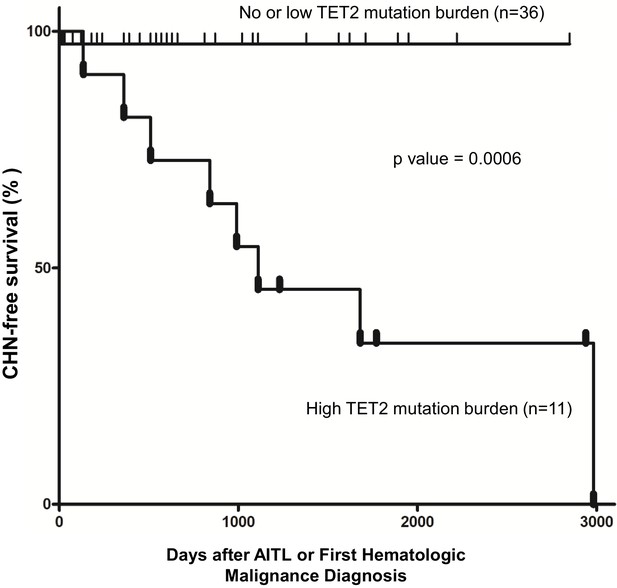
Pathogenic TET2 mutation status in the bone marrow/peripheral blood (BM/PB) samples is a predictive biomarker for concomitant hematologic neoplasms in angioimmunoblastic T-cell lymphoma (AITL) patients.
Kaplan–Meier analysis of concomitant hematologic neoplasm-free survival in AITL or AITL-related patients based on TET2 mutation status in the BM/PB. Concomitant hematologic neoplasm-free survival of AITL patients can be stratified based on absent/low or high TET2 mutation burden subgroups. p-Value was calculated by log-rank test, and p-value<0.05 is considered statistically significant. In one case, the second hematologic malignancy (PV) preceded the development of AITL.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Commercial assay or kit | KAPA HyperPlus Kit | Roche | Catalog # 07962363001 | |
| Commercial assay or kit | Twist Hybridization and Wash Kit | Twist Bioscience | Catalog #101025 | |
| Commercial assay or kit | Lymphoma pilot (16X), lot 3020 | Twist Bioscience | Catalog #3020 | |
| Commercial assay or kit | HiSeq 3000/4000 SBS Kit | Illumina | Catalog # FC-410-1001 | |
| Commercial assay or kit | HiSeq 3000/4000 PE Cluster Kit | Illumina | Catalog #PE-410-1001 | |
| Other | HiSeq 4000 System | Illumina | RRID:SCR_016386 | |
| Software, algorithm | NextGENe | SoftGenetics, LLC | RRID:SCR_011859 Catalog # NG001 version 2.4.2.3 | |
| Software, algorithm | MutSignature | Fantini, 2021, https://github.com/dami82/mutSignatures | Version 2.1.1 | |
| Software, algorithm | Maftools | Mayakonda, 2021, https://github.com/PoisonAlien/maftools | Version 2.4.12 | |
| Software, algorithm | R base package | https://www.r-project.org/ | RRID:SCR_002394version 4.0.2 | |
| Software, algorithm | Prism | GraphPad | RRID:SCR_002798version 5 |
Additional files
-
Supplementary file 1
Clinicopathological information of patients.
AITL: angioimmunoblastic T-cell lymphoma; BM: bone marrow; CH: clonal hematopoiesis; CMML: chronic myelomonocytic leukemia; CTCL: cutaneous T-cell lymphoma; DLBCL: diffuse large B-cell lymphoma; Dx: diagnosis; LN: lymph node; MCF: multi-color flow cytometry; MF: myelofibrosis; MPN: myeloproliferative neoplasm; n/a: data unavailable; PB: peripheral blood; PTCL: peripheral T-cell lymphoma; PV: polycythemia vera; T: type of specimen; TB: estimated neoplastic T-cell burden in BM. &Interval (days) between the lymph node and BM sampling time: negative numbers represent days before diagnosis of AITL or PTCL-NOS by the lymph node biopsy; positive numbers mean days after diagnosis of AITL or PTCL-NOS by the lymph node biopsy. $This TB was estimated based on the limit of detection of the TCRG assay, which is 1–5%.
- https://cdn.elifesciences.org/articles/66395/elife-66395-supp1-v2.xlsx
-
Supplementary file 2
The T-cell and myeloid next-generation sequencing (NGS) targeted panels.
The gene list shows the genes covered by the T-cell and myeloid targeted panels, respectively. The sequencing summary spreadsheet compares the sequencing performance of the T-cell targeted panel and the myeloid targeted panel in five bone marrow/peripheral blood (BM/PB) samples.
- https://cdn.elifesciences.org/articles/66395/elife-66395-supp2-v2.xlsx
-
Supplementary file 3
Summary of the variants identified by the T-cell targeted panel in angioimmunoblastic T-cell lymphoma/peripheral T-cell lymphoma, not otherwise specified (AITL/PTCL-NOS) and matched bone marrow/peripheral blood (BM/PB).
Variant allele frequencies (VAF, %). VAF values highlighted in red represent those attributable to involvement by AITL. VAF values highlighted in blue represent those of the variants present only in the BM or PB. TFH: follicular T helper cell phenotype. C to A base substitution is colored in green. &Detected in diffuse large B-cell lymphoma (DLBCL) (renal tissue) by the myeloid panel.
- https://cdn.elifesciences.org/articles/66395/elife-66395-supp3-v2.xlsx
-
Supplementary file 4
Summary of variant types.
Mean and median, the average and median number of mutations of the specified subtype per sample, respectively. Percentage, percentage of mutations with the specified subtype relative to the total number of mutations. *A total of 21 bone marrow and 6 PBL samples were sequenced. Among the 27 bone marrow/peripheral blood (BM/PB) samples, 20 had identifiable mutated genes after filtering out the variants due to involvement by the neoplastic T-cells.
- https://cdn.elifesciences.org/articles/66395/elife-66395-supp4-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/66395/elife-66395-transrepform1-v2.docx